Spring semester BMB 484
Instructor: Ross Hardison rch8@psu.edu 304 Wartik Lab 814-863-0113

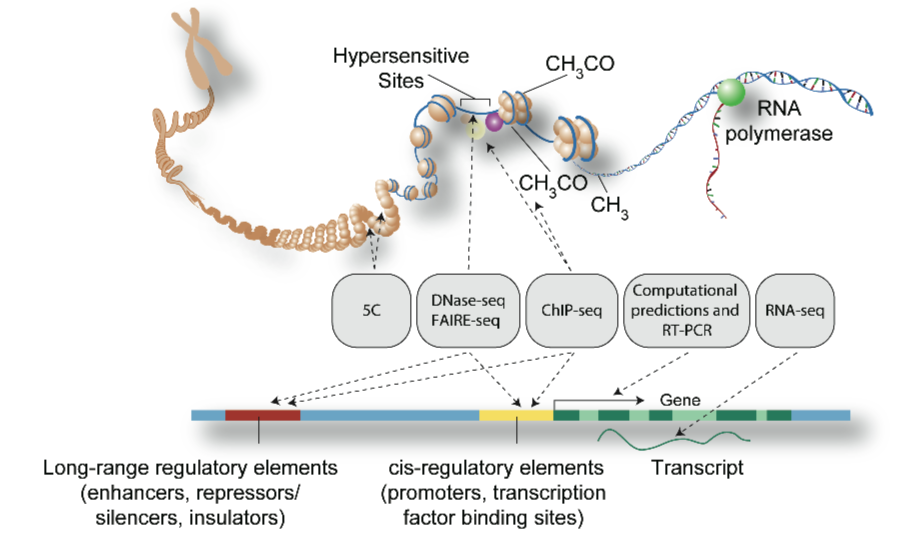
Genomic technologies enable the determination of many function-associated features at near-nucleotide resolution. Fig. 1a from A User's Guide to the Encyclopedia of DNA Elements (ENCODE) in PLoS Biology.
What is it in your DNA that makes you different from chimpanzees, mice or flies? What sequences in your DNA make it more or less likely that you will develop diabetes or cancer?
These are questions of widespread interest, answers to which could play a major role in personalized medicine and in our understanding of our place in the biosphere. Modern genomic analysis is bringing great insights to their pursuit, with occasionally some very exciting answers. One hallmark of contemporary genomic research is that data are released rapidly along with tools for browsing and analyzing it. Thus not only can you learn the major results by reading papers, but you can examine the underlying data and do your own analyses. Discovery is not longer the exclusive domain of the data producers - you can join in!
One goal of genomics, and one that affects all of the life sciences, is the quest to identify all the functional regions of a genome. But just as we cannot see most of the matter in the universe (it is "dark"), we currently know little or nothing about the function of most of the DNA in complex genomes. A full understanding of the functional regions of even the smallest, simplest genomes eludes us. Hence distinguishing functional from "neutral" DNA is a major contemporary challenge for those studying any species. The challenge becomes more difficult as the genome size increases.
The goal of this course is to introduce students to some of the ongoing research aimed at identifying functional regions in genomes and to encourage them to use web-based bioinformatics tools for exploring the databases of genomic and epigenetic data. Students will be expected to develop creative projects that address issues in functional genomics of high interest to them. The objective of each project will be determined mainly by the student's interest; it can be research or educational. The medium used to convey the results of the student's work will be chosen by the student in consultation with the instructor. Employing traditional media such as term papers or oral presentations is fine, and creative and effective use of novel media (video, music, dance, etc.) is encouraged.
This course is appropriate for upper-level undergraduates and graduate students. Students should have knowledge of molecular and cell biology and genetics, and not be reluctant to talk in class. Computer programming skills are not required, but can be used in projects.
Readings used in the course over the past few years are posted on the course web site here. (The course was initially called "Comparative Genomics.") They consist of some survey chapters and articles on comparative genomics, plus more in-depth articles from the primary literature. The papers and assignments for each year are posted at the site on PSU's ANGEL.
The students will be graded on reports from homework assignments, class participation and a project. Projects can range from summaries of current knowledge, to new research results, to new methods of presenting material.
I hope this course will also serve as a forum to explore new approaches to self- and group-learning. I am convinced that using class time only for lectures and evaluation by multiple-choice tests are not the most effective means of teaching. I expect that they will not even be supported in any course within a few years. However, I doubt that I have the most creative ideas for alternative ways to conduct classes. Thus I invite and encourage students to come up with ideas for effective interactions during the classroom time. One option for the projects is to develop creative and engaging media for learning comparative genomics. My hope is that the YouTube generation will inspire some methods that take us far beyond PowerPoint. The class size has been limited to 24 to facilitate the interactions.
Course material
Some results from Spring 2008
Some results from Spring 2009
Some results from Spring 2010
Some results from Spring 2014
Some interesting examples of creative pedagogy
Page created: Sunday, 02-Dec-2007, last modified 10-Jan-2012