Credit: Wilfried Guiblet
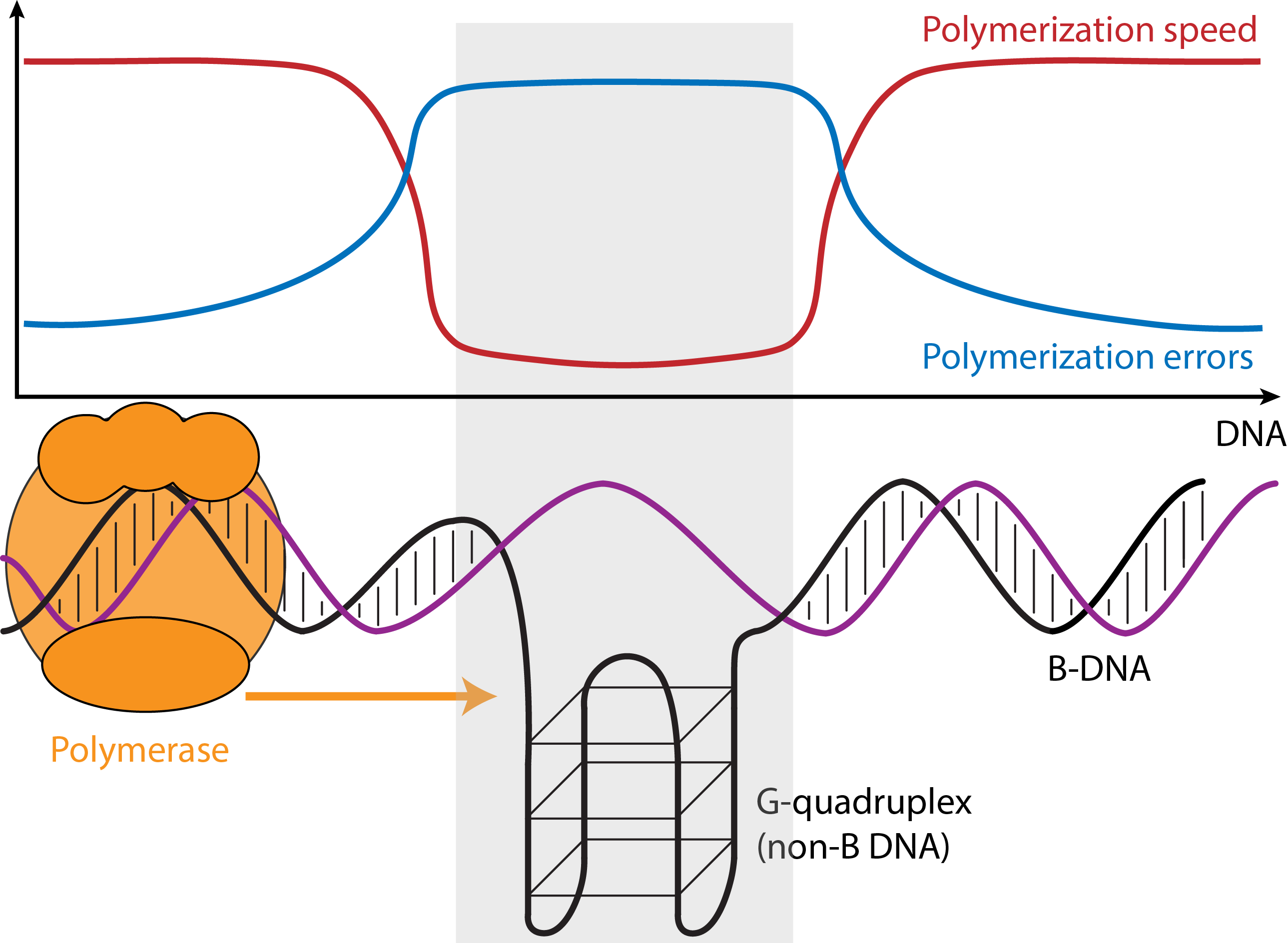
Approximately 13% of the genome can fold into DNA structures different from the B-form double-helix discovered by Watson and Crick. Such “non-B DNA” structures include cruciforms, triplexes, slipped-strand structures, G-quadruplexes (G4s), Z-DNA, and bent DNA. They form at certain DNA sequence motifs that are ubiquitous in the genomes of prokaryotes and eukaryotes. Non-B structures form intermittently and can be stabilized under certain conditions in the cell. They are emerging as key regulators of many cellular processes (e.g., replication and transcription), which may be sensitive to the environment and to temporal signals associated with cell cycle and development. Non-B DNA also affects mutation rates. Despite their clear importance in the genome, non-B DNA motifs have been critically underexplored, and studying their evolution will bring us closer to deciphering their functions in the cell. Note that many microsatellites form non-B DNA.
Collaborators: Kristin Eckert and Francesca Chiaromonte








