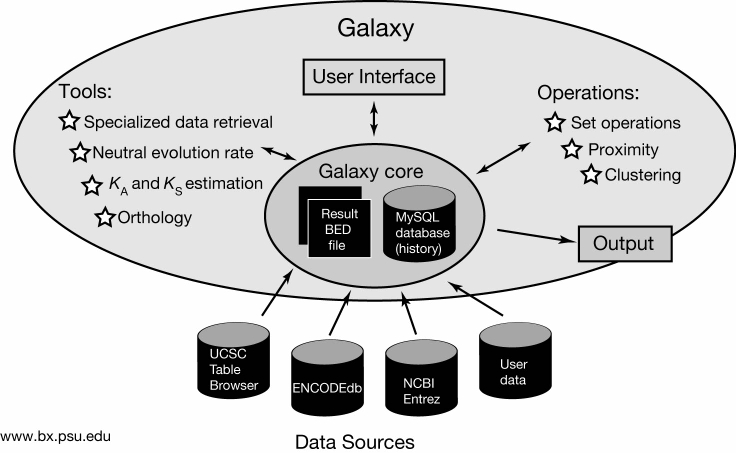
Galaxy is a metaserver that integrates various types of genomic data (sequences, alignments, microarray data) with analysis tools in a single powerful interface. Instead of being a stand-alone database, it queries remote data sources (e.g., UCSC Table Browser, ENCODEdb) to retrieve information that it then maintains, displays, and analyzes. Users' results are stored in a History page where they can be viewed, combined, filtered, and used as input for the analysis tools. Results can also be downloaded in various data formats for further processing on the user's own machine or by other web resources.
The Galaxy project is a collaborative effort involving members of the Center for Comparative Genomics and Bioinformatics at Penn State University, the University of California at Santa Cruz Genome Bioinformatics Group, the NHGRI ENCODE consortium, and NCBI.
Schematic diagram showing major components and information flow:

Poster at the 2005 CSHL Biology of Genomes meeting: PDF [629 Kb]