B M B 400
Part Four: Gene Regulation
Section V = Chapter 19
REGULATION OF EUKARYOTIC GENES
A. Promoters
1. Eukaryotic genes differ in their
state of expression
a. Recall from Part One of the course that most genes in eukaryotes are not expressed in any given tissue.
Of the approximately 30,000 genes in humans, any particular tissue will express a few at high abundance (these are frequently tissue specific, e.g. globin genes in red cells) and up to a few thousand at low abundance (these frequently encode functions needed in all cells, i.e. "housekeeping genes." You can measure this by the kinetics of hybridization between mRNA and cDNA.
b. The genes that are not expressed are frequently in an "inactive" region of the chromatin. The basic model is that genes that will not be expressed are kept in a default "off" state because they are packaged into a conformation of chromatin that prevents expression.
c. Expression of a gene then requires opening of a chromatin domain, followed by the steps discussed in Part Three of this course: assembly of a transcription complex. transcription, splicing and other processing events, translation, and any requisite post-translational modifications.
d. Various active genes can be transcribed at distinctive rates, primarily determined by the differences in rate of initiation. This ultimately produces the characteristic abundance of each mRNA, ranging from very high to very low.
2. Those genes that are expressed can be transcribed at a basal rate from the "basal” or “minimal” promoter, and in many cases they also can be induced to a high level of expression.
The process of going from no expression to basal expression may differ fundamentally from the process of going from basal expression to activated high-level expression. For instance, for some genes the former could require that the strong negative effect of silencing chromatin be removed, whereas the latter could involve covalent modification of particular transcriptional activators. However, the full mechanistic details of both processes are not yet known, although it is clear that several enzymatic activities, many of them composed of multiple polypeptide subunits, are involved in each. Changes in chromatin structure and roles for transcriptional activators have been proposed in both processes, so in fact there may be more similarity than one would have supposed initially. The fact is that we simply do not know at this time. Adding complexity to ambiguity, one should realize that the mechanisms may differ among the many genes in an organism.
Both processes (going from no expression to basal expression, and going from basal to activated expression) are part of transcriptional activation, which is currently an area of intense investigation in molecular genetics. Thus, even though a full understanding of this process eludes us, it is important to explore what is currently known about gene regulation in eukaryotes, as well as some of the still-unanswered questions. That is what we will do in Chapters 19 and 20.
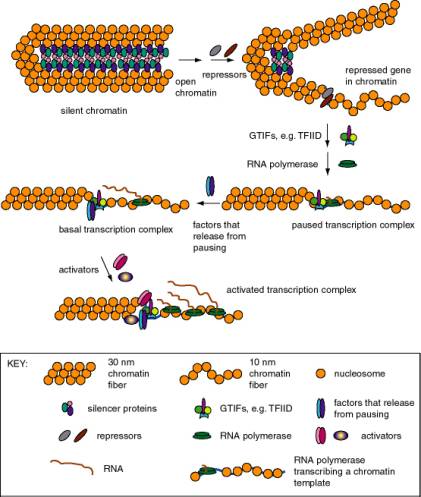
Figure 4.5.1. Expression states of promoters for RNA polymerase II. Each of these states has been described for particular genes, but it is not clear that all states are in one obligatory pathway. For instance, it possible that some gene activation events could go from silent chromatin to basal transcription without passing through open but repressed and paused transcription.
a. Basal transcription
(1) Is frequently studied by in vitro transcription, using defined templates and either extracts from nuclei or purified components.
(2) Requires RNA polymerase with general transcription factors (e.g. TFIID, TFIIA, TFIIB, TFIIE, TFIIF, and TFIIH for RNA polymerase II), as previously covered in Part Three.
b. Activated transcription
(1) Occurs via transcriptional activators interacting directly or indirectly with the general transcription complex to increase the efficiency of initiation.
(2) The transcriptional activators may bind to specific DNA sequences in the upstream promoter elements, or they may bind to enhancers (see Section B below).
(3) The basic idea is to increase the local concentration of the general transcription factors so the initiation complex can be assembled more readily. The fact that the activators are bound to DNA that is close to the target (or becomes close because of looping of the DNA) means that the local concentration of that protein is high, and hence it can boost the local concentration of the interacting general transcription factors.
3. Stalled polymerases
a. RNA polymerase will transcribe about 20 to 40 nucleotides at the start of some genes and then stall at a pause site. The classic example are heat-shock genes in Drosophila, but other cases are also known.
b. These genes are activated by release of stalled polymerases to elongate. In the case of the heat shock genes, this requires heat shock transcription factor (HSTF). The mechanism is still under study; some interesting ideas are
(1) Phosphorylation of the CTD of the large subunit of RNA polymerase II causes release to elongation ("promoter clearance"). One candidate (but not the only one) for the CTD kinase is TFIIH.
(2) Addition of a processivity factor (analogous to E. coli Nus A?), maybe TFIIS.
B. Silencers
Silencers are cis-acting regulatory sequences that reduce the expression from a promoter in a manner independent of position or orientation - i.e. they have the opposite effect of an enhancer. Two examples are the silencers that prevent expression of the a or a genes at the silent loci of the mating type switching system in yeast and silencers at telomeres in yeast.
The silencers work by sequence
specific proteins, such as Rap1, binding to DNA in chromatin. These proteins serve as anchors for expansion
of repressed chromatin. They do this by recruiting silencing proteins
called SIR proteins, named for their activity as silent information
regulators. The SIR proteins assemble the chromatin into a large complex
that is not transcribed. In this complex, the H3 and H4 histones in the
nucleosomes have hypoacetylated
N-terminal tails, the DNA can be methylated, and the entire silenced complex is
resistant to DNase digestion in vitro, all of which are characteristic of condensed,
closed chromatin. The large multiprotein complex may be inaccessible to
positive transcription factors and RNA polymerase. Thus silencing is a process
of preventing gene expression by packaging the gene into heterochromatin.
Discrete DNA sequences can be mapped as silencers by assaying the effects of deleting these sequences from chromosomes in cells. Removal of a silencer leads to depression of the regulated genes.
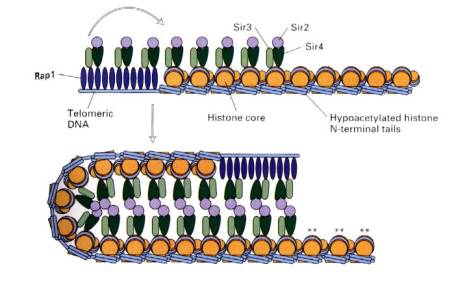
Figure 4.5.2. Transcriptionally silent chromatin, mediated by Rap1 and SIR proteins.
C. Enhancers
1. Enhancers are cis-acting regulatory sequences that increase level of expression of a gene, but they operate independently of position and orientation. These last two operational criteria distinguish enhancers from promoters.
2. Examples
a. SV40 control region
(1) SV40 (simian virus 40) infects monkey kidney cells, and it will also cause transformation of rodent cells. It has a double stranded DNA genome of about 5 kb. Because of its involvement in tumorigenesis, it has been a favorite subject of molecular virologists. The early region encodes tumor antigens (T-Ag and t-Ag) with many functions, including stimulating DNA replication of SV40 and blocking the action of endogenous tumor suppressors like p53 (the 1993 "Molecule of the Year"). The late region encodes three capsid proteins called VP1, VP2 and VP3 (viral protein n). A region between the early and late genes controls both replication and transcription of both classes of genes.
(2) The control region has an origin of replication with binding sites for T-Ag.
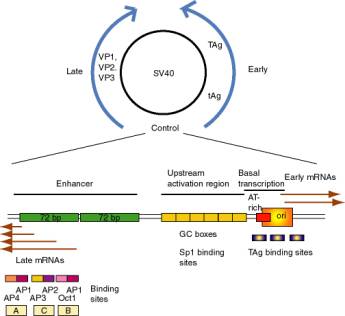
Figure 4.5.3. SV40 and its control region.
(3) Transcription initiation sites for early genes overlap the origin. Upstream from the initiation sites is an A+T rich region analogous to the TATA box, which is the binding site for TFIID. Immediately upstream are three copies of a 21 bp sequence. Each 21 bp repeat has two "GC" boxes which are binding sites for the transcriptional activator Sp1.
(a) The initiation sites + AT rich region + 6 GC boxes can be considered the promoter for early gene transcription in SV40.
(b) The consensus GC box is GGGCGG (or its complement CCGCCC). A high affinity site is GGGGCGGGG.
(4) Upstream from the early promoter are two repeats of 72 bp that comprise the enhancer.
(a) One copy of the 72 bp region has three domains that function in enhancement, called A, C and B.
(b) Each domain has binding sites for two activator proteins encoded by the host cell.
Domain B has sites for Oct1 and AP1 (Activator Protein 1, a family of proteins that includes the Jun-Fos heterodimer).
Domain C has sites for AP2 and AP3 (a protein that binds to CAC motifs in DNA).
Domain A has sites for AP1 and AP4.
(5) The enhancer was discovered by studying the effects of mutations in SV40.
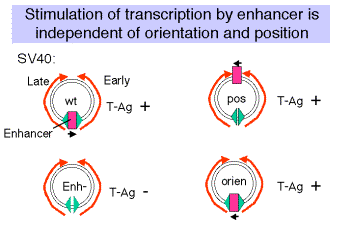
Figure 4.5.4
(a) Wild type SV40 expresses T-Ag upon infection of monkey cells and lyses infected cells. However, a viral strain lacking the 72 bp repeats shows a highly reduced level of T-Ag and rarely lyses infected cells.
(b) If the 72 bp repeats are added back to the mutant SV40 genome, except they are placed between the ends of the early and late genes (180° from their wild-type position), T-Ag is expressed at a high level and one obtains productive infections.
(c) If the orientation of the 72 bp repeats is reversed, one still gets high level expression of viral genes and productive infection. In fact, it is needed for expression of the late genes in the wild-type, which are transcribed in the opposite direction from the early genes.
(d) One concludes that the enhancer is needed for efficient transcription of the target promoters, but it can act in either orientation and at a variety of different positions and distances from the targets.
(e) Work done virtually concurrently with that described above showed that the 72 bp repeats work on other "heterologous" genes, so that, for example b-globin genes could be expressed in nonerythroid cells. In fact this was one of the key observations in the discovery of the enhancer.
(f) One copy of the 72 bp region will work as an enhancer, but two copies work better.
b. Immunoglobulin genes
(1) This was the first enhancer of a cellular gene discovered. Researchers noted that a region of the intron was exceptionally well conserved among human, rabbit and mouse sequences, and subsequent deletion experiments showed that the intronic enhancer was required for expression.
(2) After rearrangement of the immunoglobulin gene to fuse VDJ regions, one is left with a large intron between this combined variable region gene and the constant region. An enhancer is found in that intron, and another enhancer is found 3' to the polyA addition site.
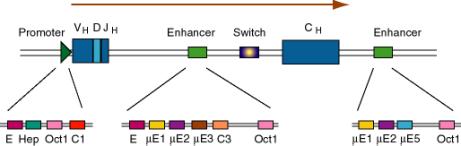
Figure 4.5.5. Enhancers in the intron and 3’ flank of an immunoglobulin gene.
(3) The enhancers have multiple binding sites for transcriptional regulatory proteins
(a) Several of these sites are named for the enhancer they were discovered in. E.g. mE1, mE2, etc. are binding sites for enhancer proteins identified in the gene for the immunoglobulin heavy chain m (mu).
The protein YY1 (ying yang 1) binds to the mE1 site (CCAT is the core of the consensus) and bends DNA there.
The octamer site (ATTTGCAT) is bound by two related proteins. Oct1 is found in all tissues examined, whereas Oct2 is lymphoid specific - the first example of a tissue-specific transcription factor. Transcriptional activators that do not have their own DNA binding sequence, like VP16 from Herpes virus, will bind to Oct proteins, which bind to DNA, and the complex can activate transcription.
(b) Some proteins will bind to sites both in the promoter and the enhancer, e.g. Oct proteins. Remember Oct1 also acts at the SV40 enhancer.
c. Summary
(1) The position of the enhancer can be virtually anywhere relative to the gene, but the promoter is always at the 5' end.
(2) Examples are known of enhancers 5' to the gene (upstream), adjacent to the promoter (like in SV40), downstream from the gene (some globin genes), within the gene (immunoglobulins) or far upstream within a locus control region (globin genes, see Chapter 20.)
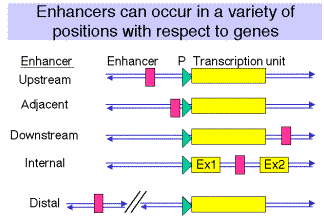
Figure
4.5.6.
3. Multiple binding sites for
transcriptional activators
a. All enhancers characterized thus far have multiple binding sites for activator proteins.
b. Multiples of binding sites are needed for function of the enhancer.
(1) In experiments with the SV40 enhancer, it was noted that some mutations that decreased the infectivity of the virus caused a mutation of one of the domains of the enhancer, e.g. domain A. When these mutants were then selected for pseudo-revertants to wild-type, with infectivity largely restored, it was found that the pseudo-revertants had duplicated one of the remaining domains. Subsequently, multimers of the various protein-binding sites were shown to be active, but monomers had little activity.
(2) The domain (e.g. A, C and B in the SV40 enhancer) with at least two binding sites is called an enhanson. Multiple enhansons make up an enhancer.
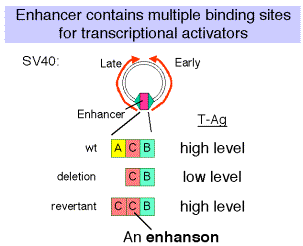
Figure
4.5.7.
C. Activator proteins and other regulators
1. Modular construction
a. DNA binding domain: Sequence-specific, direct contact with DNA
b. Multimerization domain: Allows formation of homo- or heter-multimers
c. Activation domain: direct or indirect interaction with targets (directly or directly affecting the efficiency of transcription).
2. Example: GAL4
a. After induction with galactose, the GAL4 protein will stimulate expression of genes in the GAL regulon of yeast, which encodes the enzymes that catalyze entry of galactose into intermediary metabolism. E.g. GAL1 encodes galactokinase, which converts the substrate to galactose-1-phosphate. GAL 80 keeps the regulon off in the absence of galactose.
b. The first 100 amino acids comprise the DNA binding domain of GAL4. A dimer of GAL4 protein binds to a 17 bp sequence with dyad symmetry called UASG, for upstream activating sequence for the galactose regulon.
c. The dimerization domain overlaps the DNA binding domain, encompassing amino acids 65 to 98.
d. The principle activation domain is an acidic region at the C terminus.

Figure 4.5.8. Modular structure of GAL4 protein.
e. Negative regulation is achieved by GAL80 binding at the C terminus and essentially hiding the activation domain. When induced by galactose, the GAL80 protein is altered and the activation domain is exposed. Induction causes the GAL80 protein to either dissociate or to move to a different position on GAL4 so that the activation domain is exposed.
f. Another activation domain from amino acids 148 to 196 is active in vitro, but may not be very important in the yeast cell.
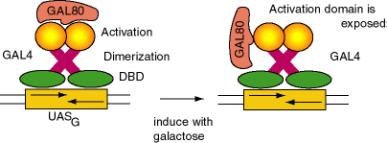
Figure
4.5.9. Negative regulation and induction of GAL4
3. Functional domains are
interchangable: "Domain swap" experiments
(1) Replacement of the DNA binding domain with a different one will change the site at which the activator will act, but not affect its ability to activate a target promoter. In other words, the DNA binding domain can be altered without affecting the activation domain, and vice versa.
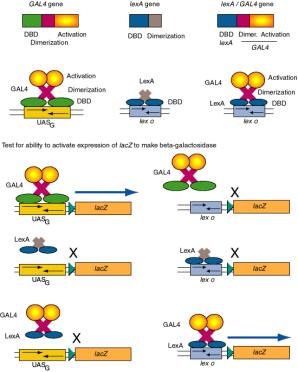
Figure 4.5.10. Domain swap experiments show DNA binding domains and activation domains are interchangable.
(2) Consider the ability of GAL4 protein to activate the promoter of the GAL1 gene.
The GAL1 promoter has a binding site for GAL4 (UASG), and in the presence of galactose, GAL4 will activate its expression. If the UASG is replaced by the operator for LexA (the repressor that regulates SOS functions in E. coli - recall this from Part Two), then GAL4 protein will no longer activate the modified GAL1 promoter. However, a hybrid protein with the DNA binding domain of LexA and the activation domain of the GAL4 protein will activate the modified promoter with the LexA operator. Similar domain swap experiments are widely used to identify functional domains of regulatory proteins.
(e) This same principle is applied in the “two-hybrid” system to identify cDNAs of proteins that interact with a designated protein.
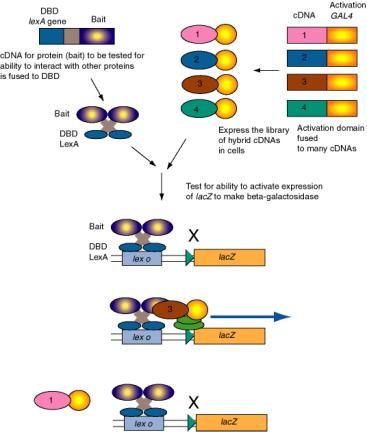
Figure 4.5.11. Two-hybrid screen for interacting proteins.
The two-hybrid screening method is a rapid and sensitive way to test a large group of proteins for their ability to interact in vivo with a particular protein. For example, one component of a regulatory complex may be characterized and a cDNA available. This cDNA for the “bait” protein is fused to a DNA segments encoding a well-known DNA binding domain, such as that of LexA, which binds to lex o. When introduced into yeast cells with the lacZ gene (encoding beta-galactosidase) under control of lex o, the lacZ gene is not expressed because the hybrid bait protein has no activation domain. A library of cDNAs to be tested are fused to the DNA encoding the activation domain of GAL2. When these are transformed into yeast cells carrying the hybrid LexA_DBD-bait and the lex o - lacZ reporter, only the hybrid proteins that interact with the bait will stimulate expression of lacZ. Transformed cells that are positive in this assay are carrying a plasmid with a hybrid gene with the cDNA encoding a protein (the “trap”) that interacts with the protein of interest (bait).
D. DNA
binding domains
Computer-assisted three-dimensional views of several transcription factors, illustrating many of the domains described here, can be viewed as Chime tutorials at
http://www.bmb.psu.edu/pugh/514/mdls
http://www.clunet.edu/BioDev/OMM/cro/cromast.htm
1. Helix-turn-helix, homeodomain
(1) The sequence of the "homeodomain" forms three helices separated by tight turns.
(2) Helix three occupies the major groove at the binding site on the DNA. It is the recognition helix, forming specific interactions (H-bonds and hydrophobic interactions) with the edges of the base pairs in the major groove.
(3) Helices one and two are perpendicular to and above helix three, providing alignment with the phosphodiester backbone. The N-terminal tail of helix interacts with the minor groove of the DNA on the opposite face of the DNA.
(4) Helix two + helix three is comparable to the helix-turn-helix motif first identified in the l Cro and repressor system.
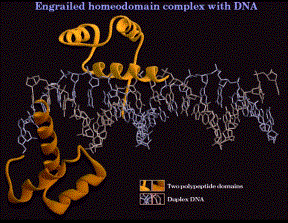
Figure 4.5.12. Helix-turn-helix in the “homeodomain”
(5) Examples
(a) Homeotic genes and their relatives.
All these are involved in regulating early developmental events in Drosophila. They are transcription factors (regulating the genes that determine the next developmental fate), and they have this same protein motif for their DNA binding domains.
Some specific examples are the products of these genes:
the pair-rule gene eve = even skipped
the segment polarity gene en = engrailed
the homeotic gene Antp = antennapedia
Recent review: Scott, M. (1994) Cell 79:1121-1124.
(b) Other proteins
Oct proteins; Oct 1 is found in all cells examined and Oct2 is lymphoid specific. Both bind to the octamer sequence. In both these proteins, the homeodomain is preceded by another important protein motif called a POU domain.
2. Zinc fingers
(1) Cys2His2
(a) Consensus sequence:
Cys-X2-4-Cys-X3-Phe-X5-Leu-X2-His-X3-His
(b) The thiol of each of the 2 Cys and one of the ring nitrogens of the imidazole of each of the 2 His donate electron pairs to form a tetrahedral coordination complex with Zn2+. This forms the base of the "finger."
(c) The "left" half of the finger (with the 2 Cys) forms two beta sheets, and the "right" half (with the 2 His) forms an a-helix. This was predicted from the expected secondary structures of this sequence, and was subsequently demonstrated by 2-D NMR and confirmed by X-ray diffraction analysis of crystals.
![]()
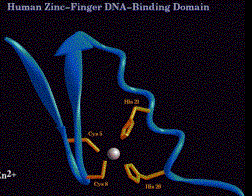
Figure 4.5.13. C2H2 Zn finger
(d) In a protein with 3 adjacent Zn fingers, e.g. Sp1 (remember this protein from the SV40 early promoter), each finger binds in the major groove to contact three adjacent base pairs. For the high affinity binding site, one finger contacts GGG, the next finger contacts GCG, and the remaining finger contacts GGG. So the three fingers curve along to contact the major groove for most of one turn of the helix.
(e) Members of this class of Zn finger proteins have multiple fingers, usually in a tandem array. Examples include TFIIIA (the motif was discovered in this protein) with 9 fingers, a CAC-binding protein (related to some extent to Sp1) with 3 fingers, and Drosophila ADR1 with 2 fingers.
(2) Cys2Cys2
(a) Consensus sequence:
Cys-X2-Cys-X1-3-Cys-X2-Cys
(b) Forms a distinctly different structure from the Cys2His2 Zn fingers.
[1] Note that the number of amino acids between the 2 "halves" of the finger (1 to 3 in this case) is much less than the 12 that separate the two halves of a Cys2His2 Zn finger.
[2] The Cys2Cys2 fingers are not interchangable with Cys2His2 Zn fingers in domain swap experiments.
[3] The proteins do not have extensive repetitions of the motif, in contrast to proteins with Cys2His2 Zn fingers.
(c) Found in steroid hormone receptors, e.g. glucocorticoid receptors
Each monomer of the receptor has two Cys2Cys2 fingers, one for DNA binding and the other for dimerization. Each monomer of the dimer binds to successive turns of the major groove to occupy the binding site (with a dyad symmetry).
(3) Cys6
(a) The DNA binding domain of GAL4 has 6 Cys in this sequence:
Cys-X2-Cys-X6-Cys-X6-Cys-X2-Cys-X6-Cys
(b) The 6 cysteines coordinate to 2 Zn2+ atoms to form a binuclear cluster.
(c) A great Chime presentation showing both the DNA-binding domain and dimerization domains for GAL4 can be seen at
http://www.umass.edu/microbio/chime/prsswc/2frmcont.htm
This movie shows many features of the protein quite clearly. For example, note that the DNA binding domain is mainly in contact with the sugar-phosphate backbone of the DNA, with very little contact with the major or minor grooves. This contrasts markedly with the interactions seen for other transcription factors discussed in this chapter.
(d) This particular Zn-coordinated cluster of cysteines has been seen in several yeast regulatory proteins, but is not very common in other organisms analyzed to date.
(4) GATA1
(a) GATA1 is a transcription factor, abundant in erythroid cells, that is required for erythroid differentiation. Binding sites for it have the consensus WGATAR (W = A or T, R = A or G). GATA1 binding sites are common in promoters, enhancers and locus control regions of erythroid genes.
(b) The DNA binding domain has Zn fingers, but with a distinctly different structure from the others discussed previously.
(c) This binding domain is found in several apparently unrelated DNA-binding proteins, including some fungal regulatory proteins.
(d) Several GATA-like proteins have 2 Zn fingers. One binds to a specific site on the DNA, the other finger interacts with other proteins.
3. Leucine zipper
(1) The leucine zipper per se is a dimerization domain. In each monomer, the zipper region forms an a helix with a leucine every 7 amino acids, i.e. every 2 turns of the helix. This produces a row of leucines along the hydrophobic face of the helix. The two monomers interact by intercalating the leucine along their hydrophobic surfaces. The two helices form a coiled coil. Other aliphatic amino acids like valine can substitute for leucine.
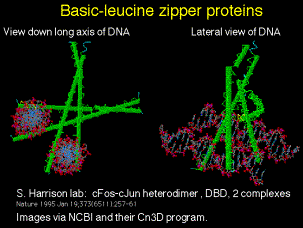
Figure 4.5.14. Basic-leucine zipper proteins
(2) A basic region is just to the N-terminal side of the leucine zipper in many proteins. The basic region plus the leucine zipper (bZip) is the DNA-binding domain.
(3) Examples
(a) C/EBP = CCAAT/enhancer binding protein, forms a homodimer
(b) AP1 family includes heterodimers of c-Fos and c-Jun
4. Basic helix-loop-helix
(1) The helix-loop-helix (HLH) motif consists of two amphipathic helices separated by a loop of variable length. An amphipathic helix simply has a hydrophobic side and a hydrophilic side. Dimerization occurs via interactions between the hydrophobic faces.
(2) The DNA binding domain consists of a basic region plus the helix-loop-helix region (bHLH).

Figure
4.5.15. Basic helix-loop-helix proteins
(3) Examples include heterodimers that can exchange partners
(a) MyoD is a key protein in committment of mesodermal tissues to muscle differentiation. Other relatives, such as myogenin and myf5, are equally important and provide redundant functions. All are muscle-specific and have a similar binding domain. MyoD is active when it has E12 or E47 as its heterodimeric partner; when active it will stimulate transcription of muscle specific genes such as the one encoding creatine kinase. E12 and E47 were initially discovered as proteins that bound to enhancers of immunoglobulin genes, but are found in virtually all cell-types. Another protein, called Id, can also bind to E12 or E47 by its HLH domain. However, Id lacks a basic domain, so heterodimers with Id are not active. So the activity of bHLH proteins can be regulated by exchange of partners.
(b) A developing theme is that one of partners of a bHLH heterodimer is ubiquitous (e.g. E12, E47 in mammals, da = daughterless in Drosophila) and the other is tissue-specific (MyoD or AC-S = achaete-scute, a regulator of neurogenesis in Drosophila). The ubiquitous components may be involved in regulating a variety of other tissue-specific proteins with bHLH domains.
(c) Myc, one of many regulators of the cell cycle, is a bHLH protein. It forms partners with Max, and it is possible that this is important in regulation of the cell cycle.
E. Transcriptional activation domains
1. Acidic
(1) This domain has been postulated to be an "acid blob" or an amphipathic helix with acidic residues on one face. Recent physico-chemical studies of GAL4 have shown b-sheet structure. At this point no single structure has been established.
(2) Examples:
GAL4 protein, VP16, GCN4, glucocorticoid hormone receptor, AP1, and the l repressor (activation of PRM).
2. Gln-rich
This domain is rich in glutamine, as its name implies. Examples of proteins containing the domain are Sp1, Antp, Oct1 and Oct2
3. Pro-rich
Again, the domain is rich in proline. Examples include CTF/NF1 (involved in regulation of replication as nuclear factor 1, and proposed to be one of many proteins binding to CCAAT motifs).
4. Work so far has not established well-defined secondary or tertiary structures for these domains. One possibility is that the activation domains assume their proper structure after binding to its target, i.e. an induced fit model.
Table 4.5.1. Selected eukarytoic transcription factors and their properties
|
Name |
System |
Binding site (top strand) |
Quaternary structure |
DNA binding domain |
Activation domain |
Other comments |
|
Engrailed |
early development |
|
|
homeodomain |
|
|
|
Sp1 |
SV40, cellular housekeeping
genes |
GGGGCGGGG |
monomer |
3 Zn fingers Cys2His2 |
Gln-rich |
phosphoprotein |
|
AP1 |
SV40, cellular enhancers |
TGASTCA |
heterodimer, Jun-Fos, Jun2, others |
basic region + Leu zipper |
acidic |
regulated by
phosphorylation |
|
Oct1 |
lymphoid and other genes |
ATTTGCAT |
monomer, but can bind VP16 |
POU domain + homeodomain
(HTH) |
Gln-rich, also binds VP16 |
Oct1 is ubiquitous, Oct2 is
lymphoid specific |
|
GAL4 |
yeast galactose regulon |
CGGASGACWGTCSTCCG |
homodimer |
Zn2Cys6, binuclear
cluster |
acidic |
|
|
Glucocorticoid receptor |
glucocorticoid responsive
genes |
TGGTACAAATGTTCT |
cytoplasm: with "heat
shock" proteins; nucleus: homodimer |
2 Zn fingers, Cys2Cys2 |
close to Zn finger |
binding of hormone ligand
changes conformation, move to nucleus and activate genes |
|
MyoD |
determination of myogenesis |
CAGCTG |
heterodimer with E12/E47:
active; heterodimer with ID:
inactive |
basic-helix-loop-helix |
|
switch partners to activate
or inactivate |
|
HMG(I)Y |
interferon gene and others |
minor groove |
monomer (?) |
|
|
bends DNA to provide
favorable interactions of other proteins |
|
VP16 |
Herpes simplex virus |
not bind tightly to DNA |
binds to proteins like Oct1 |
|
acidic activation domain;
very potent |
binds to other proteins
that themselves bind specifically to DNA |
F.
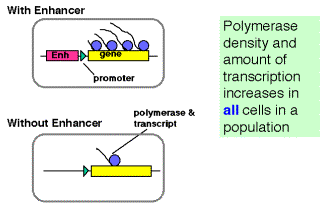
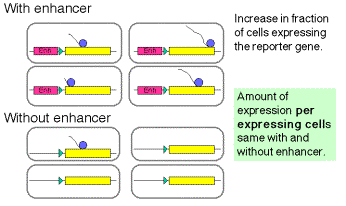
Enhancers can work by increasing the probability that a gene will be in a
transcriptionally competent region of chromatin.
Fig. 4.5.16. Some enhancers increase the rate of initiation of transcription
The latter observation implicates a chromatin-based mechanism for the mode of action of these enhancers. For instance, they may act by causing the chromatin structure to be altered around the gene, placing it in an “open” or “active” chromatin domain. Genes in “open” chromatin presumably are more accessible to the transcriptional machinery. This is discussed in more detail in Chapter 20.
G. Communication between promoter and
enhancer
1. Models: Looping vs. tracking
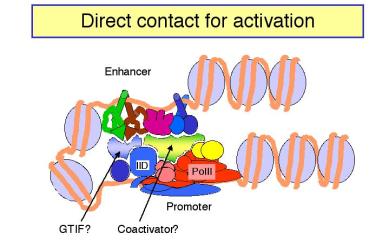
Figure 4.5.18.
a. In looping models, the activators bound to the enhancer are brought in close proximity to their targets at the promoter by forming loops in the DNA.
(1) The activators can make direct contact with their target (perhaps the pre-initiation complex), or they may operate through an intermediary called a co-activator or mediator.
(2) If a loop is formed, in principle it does not matter how large the loop is or if the activator binding site is 5' or 3' to the target. This could explain the ability of enhancers to operate independently of position.
b. In tracking models, the enhancer is an entry site for the factors that must assemble the transcription complex on the promoter. Once the activator proteins bind to the enhancer, they facilitate entry of, e.g. the general transcription factors and RNA polymerase, to the chromosome. These components then move along the chromosome until they reach the promoter, where they assemble the initiation complex. If the components can move in either direction, then an enhancer could act from any position relative to the promoter. In this model, there is no direct contact between activator proteins bound to the enhancer and the pre-initiation complex at the promoter.
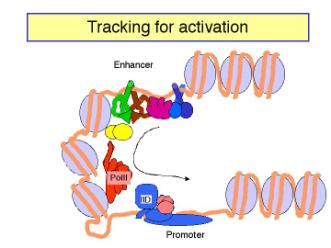
Figure 4.5.19.
c. The looping model is favored at this time. However, it has been difficult to design experiments that definitely rule out tracking. Several observations show that DNA can form loops in vitro, allowing contact between proteins at the enhancer and those at the promoter. For instance:
(1) Using electron microscopy, one can visualize loops of DNA held together by interactions between enhancer-bound activator proteins and proteins bound to the promoter.
(2) The biochemical approaches show that the activation domains of transcription factors can bind to components of the pre-initiation complex, such as TFIID (see Section H).
However, as will be discussed below, genetic experiments show that these interactions are not required for transcriptional activation of at least some genes. Even if such interactions do occur physiologically, are they part of a stable looping complex or are they transient interactions that would occur during the entry of transcription factors at the enhancer in the tracking model?
One line of evidence that has been used to support the looping model is that an enhancer located on one DNA molecule can act on a target promoter on another DNA molecule if the two molecules are held together physically by a biotin-avidin linkage. This linkage holds the DNA molecules in close proximity, which can be interpreted as mimicking a loop. However, one could also argue that tracking occurred across the biotin-avidin bridge. The ability to explain experimental results in terms of both models means that the design and available technology has not been sufficiently strong to distinguish between the two models.
Perhaps the strongest argument against the tracking model is that the physical basis for the tracking is not specified. Of course, this makes it more difficult to design experiments to test it.
As is the case for many issues in eukaryotic gene regulation, different genes may be regulated by different mechanisms.
2. Stereospecific complex: Role of proteins that bend DNA
a. Recent experiments have shown the importance of a highly specific three-dimensional nucleoprotein complex at some promoters [reviewed in Tjian and Maniatis (1994) Cell 77:5-8].
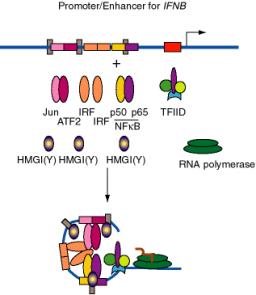
Figure 4.5.20. DNA bending by HMGI(Y) in formation of enhanceosome at IFNB promoter.
b. E.g. the enhancer for the interferon-b gene, which is located just upstream from the promoter, has binding sites for three dimeric "conventional" transcription factors: NFKB (p50 + p65), IRF, and a heterodimer of ATF2 + Jun (a relative of AP1). In addition, there are three specific binding sites for HMGI(Y).
(1) HMGI(Y) is a member of the "high mobility group" of nonhistone chromosomal proteins. Most HMG proteins are abundant in the nucleus, albeit not as abundant as histones.
(2) HMGI(Y) binds in the minor groove of DNA and bends the DNA.
(3) It also makes specific protein-protein contacts with IRF, ATF2 and NFkB, even in the absence of DNA.
(4) By bending the DNA at precise positions by a defined amount, and by aiding the binding of other proteins, HMGI(Y) seems to play a critical role in assembly of the enhancer complex in juxtaposition with the promoter.
(5) In general, proteins that bend DNA can be the agents that cause the looping to bring the enhancer-binding proteins in proximity to their targets.
c. Other proteins that bend DNA
cAMP-CAP (recall this from catabolite repression in E. coli), IHF = integration host factor (required for integration of l DNA to form a prophage, via a large complex called an intasome), and YY1 (ying yang 1) which has either negative or positive effects on a large variety of genes in mammals.
H. Targets of transcriptional activators
a. Cohesion between activation domains and targets may be driven by hydrophobic interactions.
(1) Initially it was thought that acidic activators would act on basic targets, and that Gln-rich activators would form complementary H-bonded structures with the targets.
(2) Structural work to date suggests that hydrophobic interactions interspersed with ionic bonds (acidic activators) and H-bonds (Gln-rich activators) may drive the cohesion between the two proteins.
b. Targets so far recognized are
TBP and TAFs
(1) Several strategies are used to identify targets of transcriptional activators. Two are:
(a) If one makes an affinity column with an activator domain serving as the ligand (in particular the acidic activating domain of VP16), and one pours a mixture of transcription components over it, what is bound with high specificity?
Different laboratories have equally strong data for specific binding of TBP, a protein associated with TBP in the TFIID complex, called TAFII40, and TFIIB. Many interactions have now been demonstrated to occur in vitro using similar biochemical techniques.
(b) Do mutations in these putative targets that destroy their ability to form a general transcription complex responsive to activators also prevent their binding to VP16? In all three cases, the answer is yes.
(c) Similar affinity chromatography experiments with the Gln-rich activation domain of Sp1 shows specific interaction with a different TBP-associated factor called TAFII110.
(2) Maybe acidic activators interact with all three proteins. The drawing below shows specific interaction with both TAFII40 and TFIIB, and further interactions are not out of the question. So far the best candidate for the target of Gln-rich domains is TAFII110.
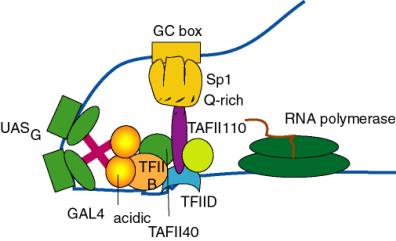
Figure 4.5.21. Targets of activation domains observed in cell-free systems.
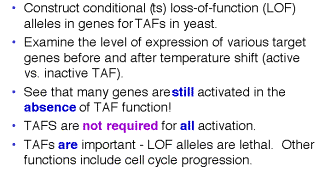
(3) However, just because a protein interacts with another one in vitro, it may not interact inside the cell. Genetic approaches are required to ascertain this. Recent studies argue that some of the TAFs implicated as targets for transcriptional activators are not required for activation of many genes, but may be for some other genes. In addition, the genetic experiments show that the TAFs have roles in other cellular processes, such as regulation of the cell cycle.
Figure
4.5.22. TAFs are not REQUIRED for
all activation
(4) The best evidence for direct contact between two proteins in vivo is to show that mutations in the gene for one of the proteins can suppress mutations in the gene for the second protein. Experiments such as these have shown conclusively that the activation domain of CAP interacts directly with the a subunit of RNA polymerase to activate certain genes in E. coli.
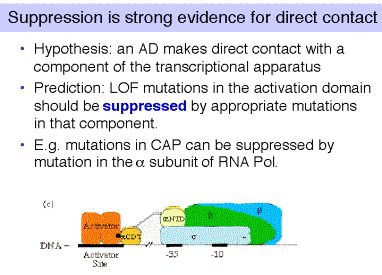
Figure 4.5.23.
I. Temporal and
tisssue-specificity via regulation of activator proteins
Four different avenues have been described in different systems.
a. The transcription factor may only be synthesized in a specific tissue, or at a particular developmental stage. Examples include homeoproteins, which are present at defined stages of development and at particular places in the embryo, and the factor GATA1 which is synthesized (almost) exclusively in erythroid and mast cells of vertebrates.
b. An inactive form of the transcription factor may be converted to an active form. For instance, phosphorylation of the heat shock transcription factor and de-phosphorylation of AP1 will activate each of these factors.
c. The active form may be imported into the nucleus after a critical conformational change. For example, binding of a steroid hormone to its receptor allows it to move to its targets in the nucleus (see next section). The protein NFkB is held in the cytoplasm in complex with IkB (inhibitor of NFkB), but when NFkB dissociates, it moves to its targets in the nucleus.
d. Exchanging partners of a heterodimer can lead to activation. For instance, when MyoD, a bHLH protein, is bound to the HLH protein Id, it is not active. But when Id is replaced by E12 or E47, it is active.
J. Induction of genes responsive to steroid hormones
The story for glucocorticoid receptor (GR) has been particularly well worked out, partly because of interest in its action on the promoter-enhancer of mouse mammary tumor virus (MMTV). In a target cell prior to exposure to the hormone, GR is complexed with a heat shock protein Hsp90 and is in an inactive conformation, with the activation domain hidden. Binding of a glucocorticoid hormone (such as cortisol or the analog dexamethasone) leads to dissociation of Hsp90 or rearrangement of the complex, coupled with a change in conformation such that the acidic activation domain is now exposed. The hormone-receptor complex then migrates into the nucleus, where dimers of the hormone-receptor complexes bind to their specific sites, called GREs, in the promoters of enhancers of responsive genes. [Dimers of the GR-hormone complex form through interactions between one of the Cys2Cys2 fingers on each monomer, and binding to the DNA is mediated through the other Cys2Cys2 finger on each monomer.] The genes are then actively transcribed, and the mRNA is exported into the cytoplasm, where hormonally-induced proteins are synthesized.
Questions for Chapter 19. Regulation of eukaryotic genes
19.1 (POB) Specific DNA binding by regulatory proteins.
A typical prokaryotic repressor protein discriminates between its specific DNA binding site (operator) and nonspecific DNA by a factor of 105 to 106. About ten molecules of the repressor per cell are sufficient to ensure a high level of repression. Assume that a very similar repressor existed in a human cell and had a similar specificity for its binding site. How many copies of the repressor would be required per cell to elicit a level of repression similar to that seen in the prokaryotic cell? (Hint: The E. coli genome contains about 4.7 million base pairs and the human haploid genome contains about 2.4 billion base pairs).
Use the following information for the next 3 problems. Let's imagine that part of the regulation of expression of the OB gene is mediated by a protein we will call OBF1. There is one binding site for OBF1 in the OB gene, and let's assume that is the only specific binding site in the haploid genome, or 2 specific sites in a diploid genome. The haploid human genome has about 3 x 109 bp, or 6 x 109 bp in a diploid genome. If we assume that about 33.3% of the nuclear DNA is in an accessible chromatin conformation, that means that about 2 x 109 bp of DNA are available to bind OBF1 nonspecifically.
19.2 The diameter of a mammalian nucleus is about 10 mm. If you model a nucleus as a sphere, what is its volume? What is the molar concentration of specific and nonspecific binding sites in the nucleus?
Binding of OBF1 to a specific site and to nonspecific sites is described by the following equations.
Let P = OBF1
Ds = a specific binding site in DNA
Dns = a nonspecific binding site in the genomic DNA
P + Ds
Ks = = 1011 M-1 (eqn 2)
Kns = = 105 M-1 (eqn 3)
19.3 What fraction of the OBF1 (or P in the equations) is not bound to either specific or nonspecific sites in the DNA?
19.4 How many molecules of OBF1 are needed per nucleus to maintain 90% occupancy of the specific sites? This condition means
= 9
Use the following information for the next seven questions.
The agouti gene in mice controls the amount and distribution of pigments within coat hairs. Some mutations of this gene also lead to adult-onset obesity, a mild diabetes-like syndrome, tumor susceptibility and recessive embryonic lethality. The gene encodes a predicted protein of 131 amino acids that has the structural features of a secreted protein, but no striking homology to other known proteins has been recognized. This protein is likely to be a regulator of melanin pigment synthesis, and it may also be a more general metabolic regulator.
Let's suppose that you are investigating the regulation of the agouti gene, and have the capacity to transfect a melanocyte cell line, which transcribes the wild-type agouti gene, and an adipocyte cell line, which transcribes the wild-type agouti gene only at a very low level. Further, you already know that the basal promoter is in a DNA segment located between -100 and +50. You make progressive 5' deletions of a fragment that includes -300 to +50, link it to a luciferase reporter gene, and transfect the constructs into melanocyte and adipocyte cells, with the following results.

19.5. What do you conclude about the region between -250 and -200?
19.6. What do you conclude about the region between -200 and -150?
19.7. What do you conclude about the region between -150 and -100?
You also investigate the binding of nuclear proteins to these DNA segments located upstream of the agouti gene. Extracts containing nuclear proteins from melanocytes were tested for the ability to bind to the fragments delineated in the deletion series above.
The fragment from -150 to -100 was used as the labeled probe in a mobility shift assay. The mobility of the free probe is shown in lane 1, and the pattern after binding to melanocyte nuclear extract is shown in lane 2. Lanes 3-14 show the mobility shifts after addition of the competitors to the binding reaction; the triangle above the lanes indicates that an increasing amount of competitor is used in successive lanes. "Self" is the same -150 to -100 fragment that is used as a probe, but it is unlabeled and present in an excess over the labeled probe (lanes 3-5). A completely different DNA (sheared E. coli DNA) was used as a nonspecific competitor (lanes 6-8). Two different duplex oligonucleotides, one containing the binding site for AP1 (lanes 9-11) and the other containing the binding site for Sp1 (lanes 12-14) were also tested. Thinner, less densely filled boxes denote bands of less intensity than the darker, thicker bands. Use these results to answer the next two questions.

19.8. What do you conclude from these data?
19.9. What sequence within the -150 to -100 segment might you expect to be bound in melanocyte nuclei?
19.10. The fragment from -200 to -150 was also used as a labeled probe in a mobility shift assay similar to that described for the -150 to -100 segment, as shown below.

What do you conclude from these data?
19.11. Some mutant alleles of the agouti gene are expressed ectopically (i.e. in the wrong tissue). Just using the information on the 5' deletions above, what region is a likely candidate for the position of a loss-of-function mutation that leads to ectopic expression in adipose tissue?
19.12 (POB) Functional domains in regulatory proteins.
A biochemist replaces the DNA‑binding domain of the yeast GAL4 protein with the DNA‑binding domain from the lambda repressor (CI) and finds that the engineered protein no longer functions as a transcriptional activator (it no longer regulates transcription of the GAL operon in yeast). What might be done to the GAL4 binding site in the DNA to make the engineered protein functional in activating GAL operon transcription?
19.13 What is the DNA-binding domain of the transcription factor Sp1?
19.14 What is the dimerization domain of the transcription factor AP1?
19.15 (ASC) Describe three mechanisms for regulating the activity of transcription factors.
19.16 (ASC) You have constructed a plasmid set containing a series of nucleotide insertions spaced along the length of the glucocorticoid-receptor gene. Each insertion encodes three or four amino acids. The map positions of the various insertions in the coding sequence of the receptor gene is as follows:
0 Glucocorticoid-receptor coding sequence 783
| |
| |
| |
Insertion: A B C D E F G H I J K L M N O P Q R S
The plasmids containing the receptor gene can be functionally expressed in CV-1 and COS cells, which contain a steroid-responsive gene. Using these cells, you determine the effect of each of these insertions in the receptor on the induction of the steroid-responsive gene and on binding of the synthetic steroid dexamethasone. The results of these analyses are summarized in the tablebelow.
Insertion Induction Dexamethasone
binding
A ++++ ++++
B ++++ ++++
C ++++ ++++
D 0 ++++
E 0 ++++
F 0 ++++
G ++++ ++++
H ++++ ++++
I + ++++
J ++++ ++++
K 0 ++++
L 0 ++++
M 0 ++++
N + ++++
O ++++ ++++
P ++++ ++++
Q 0 0
R 0 0
S 0 0
wild-type ++++ ++++
a) From this analysis, how many different functional domains does the glucocorticoid receptor have? Indicate the position of these domains relative to the insertion map.
b) Which domain is the steroid-binding domain?
c) How could you determine which of the domains is the DNA-binding domain?