B M B 400, Part
Three
Gene Expression
and Protein Synthesis
Lecture Notes
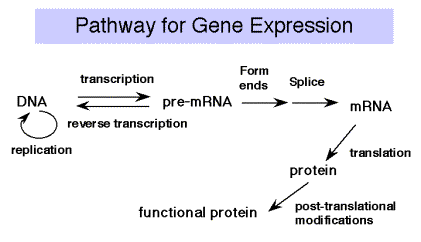
Overview of Part Three: The pathway of gene expression
Recall the Central Dogma of molecular biology:
DNA is transcribed into RNA, which is translated into protein.
We will cover the material in that order, since that is the direction that information flows. However, there are additional steps, in particular the primary transcript is frequently a precursor molecule that is processed into a mature RNA. E.g., the rRNA genes are transcribed into pre‑rRNA, that is cleaved, methylated and modified to produce mature rRNA. Many mRNAs, especially in eukarytoes, are derived from pre‑mRNAs by splicing and other processing events. This general topic will be covered after transcription and before translation.
Fig. 3.1.1
CHAPTER 10= PART THREE-I. TRANSCRIPTION: RNA polymerase
A. RNA polymerase catalyzes the DNA‑dependent
synthesis of RNA
1. RNA polymerase requires DNA as a template. In duplex DNA, the template strand of DNA is copied into RNA by RNA polymerase. The choice of nucleotides during this process is directed by base complementarity, so that the sequence of RNA synthesized is the reverse complement of the DNA template strand. It is the same sequence as the nontemplate (or top) strand, except that U's are present instead of T's.
This process of RNA synthesis directed by a DNA template, catalyzed by RNA polymerase, is called transcription.
2. RNA polymerase does not require a primer to initiate transcription.
3. RNA polymerase catalyzes the sequential addition of a ribonucleotide to the 3' end of a growing RNA chain, with the sequence of nucleotides specified by the template. The substrate NTP is added as a NMP with the liberation of pyrophosphate. This process occurs cyclically during the elongation phase of transcription.
NTP + (NMP)n n+1 + PPi
template DNA
Mg++
Fig. 3.1.2. Sequential addtion of ribonucleotides to growing RNA
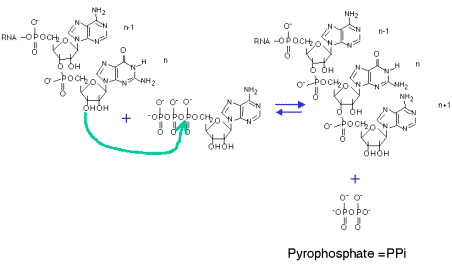
4. The liberated pyrophosphate is cleaved in the cell to 2 Pi, an energetically favorable reaction that drives the reaction in the direction of synthesis.
5. In the presence of excess PPi, the reverse reaction of pyrophosphorolysis can occur.
6. Synthesis always proceeds in a 5' to 3' direction (with respect to the growing RNA chain). The template is read in a 3' to 5' direction.
B.
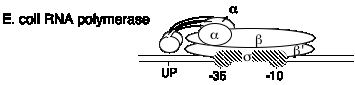
E. coli RNA polymerase structure
1. This one RNA polymerase synthesizes all classes of RNA
mRNA, rRNA, tRNA
2. It is composed of four subunits.
a. Core and holoenzyme
a2bb's a2bb' + s
Holoenzyme = a2bb's = core + s = can initiate transcription accurately as the proper site, as determined by the promoter
Core = a2bb' = can elongate a growing RNA chain
A promoter can be defined in two ways.
(a) The sequence of DNA required for accurate, specific intiation of transcription
(b) The sequence of DNA to which RNA polymerase binds to accurately initiate transcription.
b. Subunits
|
Subunit |
Size |
Gene |
Function |
|
b' |
160 kDa |
rpoC |
b' + b form the catalytic center. |
|
b |
155 kDa |
rpoB |
b' + b form the catalytic center. |
|
a |
40 kDa |
rpoA |
enzyme assembly; also binds UP sequence in the promoter |
|
s |
70 kDa (general) |
rpoD |
confers specificity for promoter; binds to -10 and -35 sites in the promoter |
Bacteria have several s factors, ranging in size from 32 to 92 kDa, each of which confers specificity for a different type of promoter.
Fig.
3.1.3. Diagram of E. coli RNA polymerase
3. Three-dimensional structure of E. coli RNA polymerase
Crystals suitable for X-ray diffraction studies have not been obtained yet, but the surface topography can be determined from by electron crystallography of two-dimensional crystalline arrays.
Fig. 3.1.4.
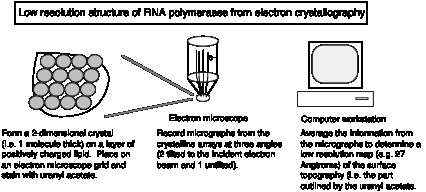
Fig. 3.1.5. E. coli RNA polymerase core (left) and holoenzyme (right)
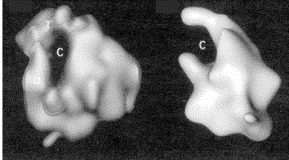
Images from analysis by Seth Darst.
The structure in the presence of s (holoenzyme) is on the right; note the open channel for DNA binding. The structure in the absence of s (the core enzyme) is on the left. Note that the channel is now closed, as if the fingers and thumbs of a hand now closed to make a circle. This striking conformational change that occurs when s dissociates is thought to confer high processivity on the RNA polymerase.
Fig. 3.1.6. Diagram of features
of E. coli RNA polymerase
holoenzyme

4. Assembly of E. coli RNA polymerase
The a subunit has two distinct domains. The N-terminal domain (a-NTD) is involved in dimerization to form a2 and further assembly of the RNA polymerase. The C-terminal domain has different functions, being used in the binding to the UP DNA sequence at promoters for rRNA and tRNA genes and in communication with many, but not all, transcriptional activators.
Fig. 3.1.7. Role of the a subunit in assembly and other functions
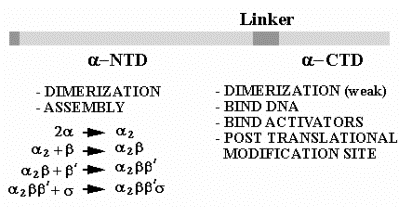
C.
E. coli RNA polymerase mechanism
1. Mode of action of s factors
The presence of the s factor causes the RNA polymerase holoenzyme to be selective in choosing the site of initiation. This is accomplished primarily through effects on the dissociation rate of RNA polymerase from DNA.
a. Core has strong affinity for general DNA sequences.
The t1/2 for dissociation of the complex of core‑DNA is about 60 min. This is useful during the elongation phase, but not during initiation.
b. Holoenzyme has a reduced affinity for general DNA; it is decreased about 104 fold.
The t1/2 for dissociation of holoenzyme from general DNA is reduced to about 1 sec.
c. Holoenzyme has a greatly increased affinity for promoter sequences.
The t1/2 for dissociation of holoenzyme from promoter sequences is of the order of hours.
2. Events at initiation of transcription
a. RNA polymerase holoenzyme binds to the promoter to form a closed complex; at this stage there is no unwinding of DNA.
b. The polymerase‑promoter complex undergoes the closed to open transition, which is a melting or unwinding of about 12 bp.
c. The initiating nucleotides can bind to the enzyme, as directed by their complementary nucleotides in the DNA template strand, and the enzyme will catalyze formation of a phosphodiester bond between them. This polymerase‑DNA‑RNA complex is referred to as the ternary complex.
d. During abortive initiation, the polymerase catalyzes synthesis of short transcripts about 6 or so nucleotides long and then releases them.
e. This phase ends when the nascent RNA of ~6 nucleotides binds to a second RNA binding site on the enzyme; this second site is distinct from the catalytic center. This binding is associated with "resetting" the catalytic center so that the enzyme will now catalyze the synthesis of oligonucleotides 7-12 long.
f. The enzyme now translocates to an new position on the template. During this process sigma leaves the complex. A conformational change in the enzyme associated with sigma leaving the complex lets the "thumb" wrap around the DNA template, locking in processivity. Thus the core enzyme catalyzes RNA synthesis during elongation, which continues until "signals" are encountered which indicate termination.
Figure
3.1.8. Events at initiation
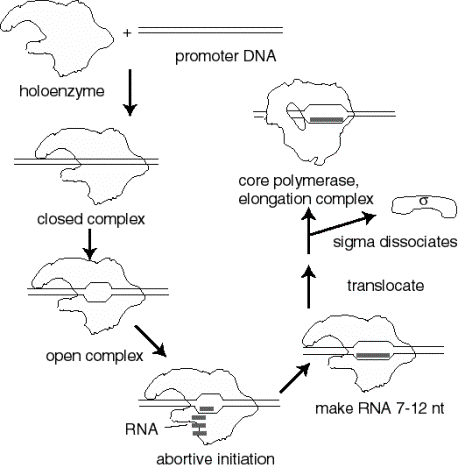
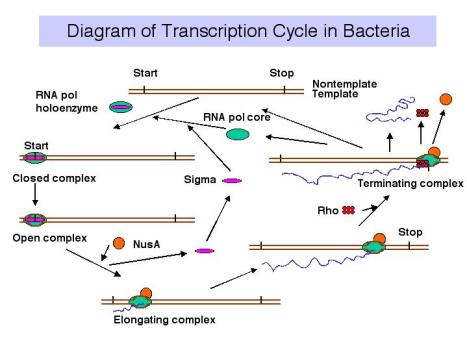
3. Transcription cycle
a. Initiation
RNA polymerase holoenzyme binds at the promoter, unwinds DNA (open complex) and form phosphodiester links between the initiating nucleotides.
b. Elongation
s dissociates and core elongates. Perhaps other factors bind to enhance the processivity (maybe NusA?)
c. Termination
At a termination signal, RNA polymerase dissociates from the DNA template and the newly synthesized RNA is released. The factor r is required at many terminators.
Figure 3.1.9.
4. Sites on RNA Polymerase core
a. The enzyme covers about 60 bp of DNA, with a transcription bubble of about 17 bp unwound.
b. The duplex DNA being transcribed is unwound at one active site on the enzyme, thereby separating the two strands (Fig. 3.1.10). The two strands are rewound at another active site, regenerating duplex DNA.
c. Within the unwound region (bubble), the 3' terminus of the growing RNA chain is bound to its complement on the template strand via H‑bonding. The DNA strand whose sequence is the same as the RNA (except for T's instead of U's) is displaced.
{This displaced strand can be called "top", "nontemplate", or "message‑synonymous strand." The template strand can also be called “bottom”, “antisense” or “message‑complementary strand.”}
Fig. 3.1.10
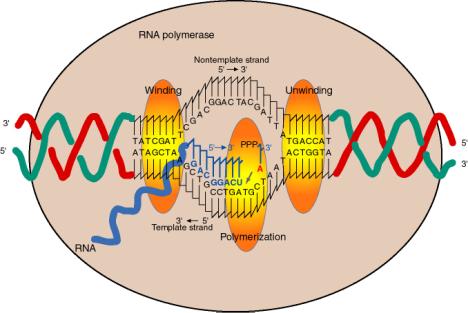
d The incoming nucleotide (NTP) that will be added to the growing RNA chain binds adjacent to the 3' end of the growing RNA chain, as directed by the template, at the active site for polymerization.
e. The incoming nucleotide is linked to the growing RNA chain by nucleophilic attack of the 3' OH on the a phosphoryl of the NTP, with liberation of pyrophosphate.
f. The reaction progresses (the enzyme moves) about 50 nts per sec. This is much slower than the rate of replication (about 1000 nts per sec).
g. If the template is topologically constrained, the DNA ahead of the RNA polymerase becomes overwound (positive superhelical turns) and the DNA behind the RNA polymerase becomes underwound (negative superhelical turns).
The effect of the unwinding of the DNA template by RNA polymerase is to decrease T by 1 for every 10 bp unwound. Thus DT = -1, and since DL = 0, then DW = +1 for every 10 bp unwound. This effect of the increase in W will be exerted in the DNA ahead of the polymerase.
The effect of rewinding the DNA template by RNA polymerase is just the opposite, of course. T will increase by 1 for every 10 bp rewound. Thus DT= +1, and since DL = 0, then DW = -1 for every 10 bp rewound. This effect of the decrease in W will be exerted in the DNA behind the polymerase, since that is where the rewinding is occurring.
5. Inhibitors: useful reagents and clues to function
a. Rifamycins, e.g. rifampicin: bind the b subunit to block initiation. The drug prevents addition of the 3rd or 4th nucleotide, hence the initiation process cannot be completed.
How do we know the site of rifampicin action is the b subunit? Mutations that confer resistance to rifampicin map to the rpoB gene.
b. Streptolydigins: bind to the b subunit to inhibit chain elongation.
These effects of rifamycins and streptolydigins, and the fact that they act on the b subunit, argue that the b subunit is required for nucleotide addition to the growing chain.
c. Heparin, a polyanion, binds to the b' subunit to prevent binding to DNA in vitro
D. Eukaryotic
RNA polymerases
1. Eukaryotes have 3 different RNA polymerases in their nuclei.
a. Each nuclear RNA polymerase is a large protein with about 8 to 14 subunits. MW is approximately 500,000 for each.
b. Each polymerase has a different function:
|
RNA polymerase |
localization |
synthesizes |
effect of a‑amanitin |
|
RNA polymerase I |
nucleolus |
pre‑rRNA |
none |
|
RNA polymerase II |
nucleoplasm |
pre‑mRNA some snRNAs |
inhibited by low concentrations (0.03 mg/ml) |
|
RNA polymerase III |
nucleoplasm |
pre‑tRNA, other small RNAs some snRNAs |
inhibited by high concentrations (100 mg/ml) |
2. Subunit structures
a. The genes and encoded proteins for the subunits of the yeast RNA polymerases have been isolated and the sequences determined, and some functional analysis has been done.
b. Some of the subunits are homologous to bacterial RNA polymerases: The largest two subunits are homologs of b and b'. The roughly 40 kDa subunit is the homolog of a.
c. Some subunits are common to all three RNA polymerases.
d. Example of yeast RNA polymerase II:
|
Approximate size (kDa) |
subunits per
polymerase |
role / comment |
|
|
220 |
1 |
related to b' |
catalytic? |
|
130 |
1 |
related to b |
catalytic? |
|
40 |
2 |
related to a |
assembly? |
|
35 |
< 1 |
|
|
|
30 |
2 |
common to all 3 |
|
|
27 |
1 |
common to all 3 |
|
|
24 |
< 1 |
|
|
|
20 |
1 |
common to all 3 |
|
|
14 |
2 |
|
|
|
10 |
1 |
|
|
|
|
|
|
|
e. The largest subunit has a carboxy‑terminal domain (CTD) with an unusual structure: tandem repeats of the sequence Tyr‑Ser‑Pro‑Thr‑Ser‑Pro‑Thr. The yeast enzyme has 26 tandem repeats and the mammalian enzyme has about 50. These can be phophorylated on Ser and Thr to give a highly charged CTD.
RNA Pol IIa is not phosphorylated in the CTD.
RNA Pol IIo is phosphorylated in the CTD.
Model: Phosphorylation of Pol IIa to make Pol IIo is needed to release the polymerase from the initiation complex and allow it to start elongation.
Figure 3.1.11.

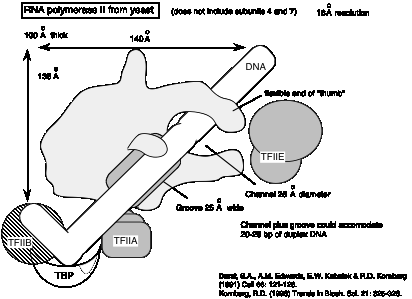
3. The 3-dimensional structure of the yeast RNA polymerase II is similar to that of RNA polymerase from E. coli.
Fig. 3.1.12
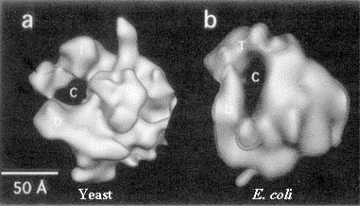
Fig.
3.1.13 Diagram of yeast RNA polymerase II with some general transcription
factors
4. RNA polymerases in chloroplasts (plastids) and mitochondria
a. The RNA polymerase found in plastids is encoded on the plastid chromosome. In some species the mitochondrial RNA polymerase is encoded by the mitochondrial DNA.
b. These organellar RNA polymerases are much more related to the bacterial RNA polymerases than to the nuclear RNA polymerases. This is a strong argument in favor of the origins of these organelles being bacterial, supporting the endo-symbiotnt model for acquisition of these organelles in eukaryotes.
c. These RNA polymerases catalyze specific transcription of organellar genes.
E. General transcription factors for eukaryotic RNA polymerase II
1. Definition
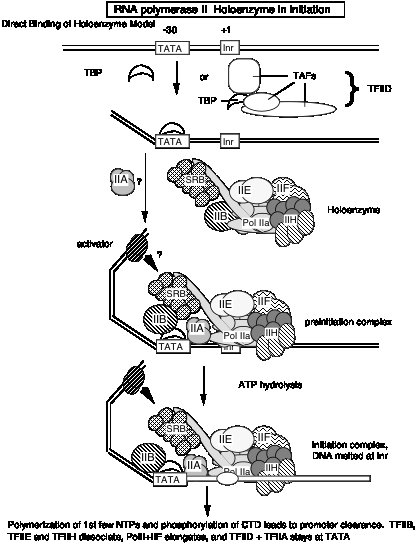
a. The general transcription factors (GTFs) are proteins required for accurate and efficient transcription that are not subunits of purified RNA polymerase. We will focus primarily on the general transcription initiation factors (GTIFs), which are proteins needed for accurate initiation of transcription. They are required for RNA polymerase to bind avidly and specifically to normal sites for transcription initiation, thereby generating specific transcripts of genes (see Fig. 3.1.14). Other transcription factors are needed for elongation.
In living cells, RNA polymerases usually start transcription at the beginning of genes. The segment of DNA required for specific initiation of transcription by RNA polymerase is called a promoter; it is commonly adjacent to the 5’ end of a gene. (Promoters will be covered in more detail in the next chapter).
Purified preparations of eukaryotic RNA polymerases can transcribe a DNA template containing a promoter, but not with specificity. The purified polymerase starts at many different sites on the DNA template, not just at the promoter. Thus some factors required for specific initiation are missing from purified eukaryotic polymerases. These specificity factors are present in crude nuclear extracts, because when such crude extracts were added to the purified polymerases, specific initiation at promoters was observed.
Biochemists purified several transcription initiation factors by fractionating nuclear extracts and assaying for this ability to confer specificity on the RNA polymerase. Several different general transcription initiation factors have been defined for each of the three eukayotic RNA polymerases.
b. The GTFs for RNA polymerase II are named TFIIx, where x = A, B, D, E, F, H, etc. These originally designated a particular chromatographic fraction that is required for accurate in vitro transcription, and now the active protein components of each fraction have been purified. TFII stands for transcription factors for RNA Pol II. TheGTFs are for RNA polymerase III are called TFIIIA, TFIIIB and TFIIIC.
Fig.
3.1.14. General transcription
factors for RNA polymerase II.
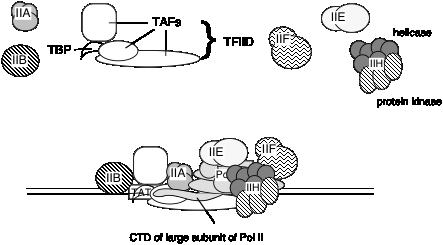
2. TFIID is a complex of many subunits. It includes the protein that binds specifically to the TATA box, called TATA binding protein = TBP, plus several TBP‑associated factors, or TAFs.
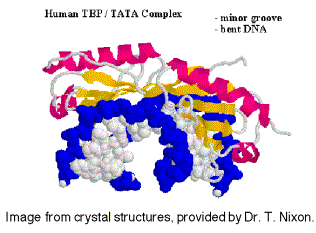
TBP binds in the narrow groove (minor grove) of DNA at the TATA box, and bends the DNA.
Fig.
3.1.15. Ribbon diagram of TBP bound to DNA.
It is not known if the same set of TAFs are in the TFIID for all promoters transcribed by RNA polymerase II, or if some are used only for certain types of promoters. TFIID is the only sequence‑specific general transcription factor so far characterized, and it binds in the minor groove of the DNA. It is also used at TATA‑less promoters, so the role of the sequence ‑specific binding is still under investigation.
3. Summary of general transcription factors for RNA polymerase II.
Factors for RNA polymerase II (human cells)
|
Factor |
No. of subunits |
Molecular mass (kDa) |
Functions |
Functions to Recruit: |
|
TFIID: TBP |
1 |
38 |
Recognize core promoter (TATA) |
TFIIB |
|
TFIID: TAFs |
12 |
15-250 |
Recognize core promoter (non-TATA); Positive and negative regulation |
RNA Pol II? |
|
TFIIA |
2 |
12, 19, 35 |
Stabilize TBP-DNA binding; Anti-repression |
|
|
TFIIB |
1 |
35 |
Select start site for RNA Pol II |
RNA PolII-TFIIF |
|
RNA Pol II |
12 |
10-220 |
Catalyze RNA synthesis |
TFIIE |
|
TFIIF |
2 |
30, 74 |
Target RNA PolII to promoter; destabilize non-specific interactions between PolII and DNA |
|
|
TFIIE |
2 |
34, 57 |
Modulate TFIIH helicase, ATPase and kinase activities; Directly enhance promoter melting? |
TFIIH |
|
TFIIH |
9 |
35-89 |
Helicase to melt promoter; CTD kinase; promoter clearance? |
|
Roeder, R.G. (1996) TIBS 21: 327-335.
4. TFIIH is a multisubunit transcription factor also involved in DNA repair.
Subunits of the human factor
|
Gene |
Molec. mass of protein (kDa) |
Function/ Structure |
Proposed Role |
|
XPB |
89 |
helicase, tracks 3' to 5' |
Unwind duplex for transcription/ Repair |
|
XPD |
80 |
helicase, tracks 5' to 3' |
Unwind duplex, Repair |
|
P62 |
62 |
unknown |
|
|
P52 |
52 |
unknown |
|
|
P44 |
44 |
Zn-finger |
Binds DNA |
|
P34 |
34 |
Zn-finger |
|
|
MAT1 |
32 |
CDK assembly factor |
|
|
Cyclin H |
38 |
Cyclin partner for CDK7/MO15 |
|
|
CDK7/MO15 |
32 |
Protein kinase |
Kinase for CTD |
Svejstrup, J.Q., P. Vichi & J.-M. Egly (1996) TIBS 21: 346-350.
TFIIH is a kinase that can phosphorylate the CTD of the large subunit of RNA polymerase II (to form Pol IIo). This step may be required to release PolII from the initiation complex so that it will begin elongation.
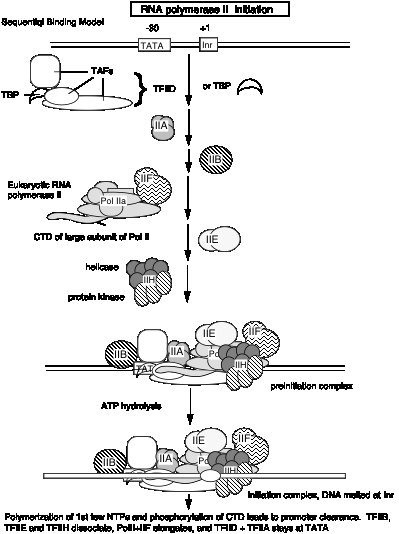
5. The general transcription factors and RNA Pol II can be assembled progressively into a preinitiation transcription complex in vitro.
Experiments using purified GTIFs and RNA polymerase II examined the ability of these proteins to assemble a specific, active complex on a particular DNA segment containing a promoter and template for transcription. The complex was formed most efficiently by adding the GTIFs and polymerase in the order shown in Fig. 3.1.16.
The complex of proteins and DNA
could be demonstrated to be specific and active because when NTPs were added,
specific transcription from the promoter was observed. We call the assembled
protein-DNA complex that is capable of specific initiation of transcription at
a promoter a preinitiation complex. As
indicated in Fig. 3.1.16, the preinitiation complex has the polymerase and
GTIFs assembled on the promoter and template. The DNA is still a duplex. An
early step in initiation is melting of the duplex at the start site for
transcription. The complex in which this has occurred can be called an
activated preinitiation complex.
Once the polymerase has begun catalyzing phosphodiester bond formation, then
the complex is an initiation complex.
The experiments showing stepwise formation of a preinitiation complex in vitro have led to the notion that binding of several of the general transcription initiation factors to DNA establishes the structure that the RNA Pol II + TFIIF complex will bind, thereby establishing the initiation site for transcription.
According to this model, the transcription factors bind to DNA in a preferred order:
TFIID, then TFIIA, then TFIIB, then RNA Pol II + TFIIF, then TFIIE
These and other factors are still being characterized. Binding of earlier factors may assist in the binding of later factors. E.g. TFIIE aids in binding of TFIIH. (See Maxon, Goodrich, Tjian (1994) G&D 8:515-524.
Fig. 3.1.16
![]()
6. RNA polymerase II holoenzyme contains the classic RNA polymerase II, some general transcription factors, and other transcriptional regulators.
Genetic analysis, largely in yeast, has shown that many other proteins in addition to RNA polymerase II and GTFs are involved in regulated transcription. Some were discovered by effects of mutations that alter regulation of genes in one or a few metabolic pathways. For instance, Gal11 is needed for regulation of the GAL operon, encoding enzymes needed for breakdown and utilization of the disaccharide galactose. Rgr1 is required for resistance to glucose repression. Note that these are the minimal roles for these proteins; they were discovered by their roles in these pathways, but could be involved in others as well.
Another class of transcriptional regulatory proteins was isolated as suppressors of alterations in RNA polymerase. Yeast strains carrying truncations in the CTD of the large subunit of RNA polymerase II fail to grow at low temperature; this is a cold-sensitive phenotype. Mutation of some other genes can restore the ability to grow at low temperature. These second site mutations that restore the wild phenotype are called suppressor mutations. Proteins identified by the ability of mutations in their genes to suppress the cold-sensitive phenotype of CTD truncations are called Srb proteins, since they are suppressors of mutations in RNA polymerase B. The ability of mutations in Srb proteins to compensate for the effects of altering RNA polymerase argues that the Srb proteins are associated with RNA polymerase in a functional complex, and this has been verified biochemically (Hengartner et al., 1995, Genes & Devel. 9:897-910).
RNA polymerase II, the GTFs, SRB proteins and other regulatory proteins have now been shown to interact in large complexes in the nucleus (Table 3.1.6). A complex called the mediator was isolated as a nuclear component needed for a response to activator proteins. Assays for in vitro transcription of DNA using purified RNA polymerase II and GTFs failed to increase the amount of transcription when transcriptional activators were added. However, a component in nuclear extracts would confer the ability to respond; this was called the mediator of activation. When purified, it was discovered to contain several Srb proteins, Gal11, Rgr1 and other transcriptional regulators.
In a separate line of investigation, an RNA polymerase II holoenzyme was discovered by isolating the complexes containing Srb proteins. This complex contains RNA polymerase II and GTFs (unlike mediator) plus many of the same proteins found in mediator, such as Srbs, Rgr1 and Gal11. This complex was shown to direct correct initiation of transcription in the presence of TBP (or TFIID) and to be capable of responding to transcriptional activators (Fig. 3.1.17).
Table 3.1.6. RNA polymerase II holoenzyme and mediator
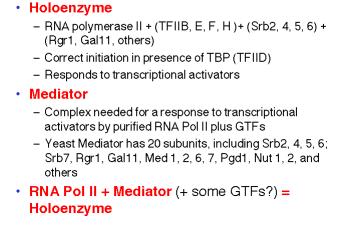
These studies show that RNA polymerase II can exist in several different states or complexes. One is in a very large holocomplex containing the mediator. In this state, it will accurately initiate transcription when directed by TFIID, and respond to activators (Table 3.1.6). The mediator subcomplex appears to be able to dissociate and reassociate with RNA polymerase II and GTFs. Indeed, this reassociation could be the step that was assayed in the identification of mediator. Without mediator, RNA polymerase II plus GTFs can initiate transcription at the correct place (as directed by TFIID), but they do not respond to activators. In the absence of GTFs, RNA polymerase II is capable of transcribing DNA templates, but it will not begin transcription at the correct site. Hence it is competent for elongation but not initiation.
Table 3.1.7. Expanding the functions of RNA polymerase II
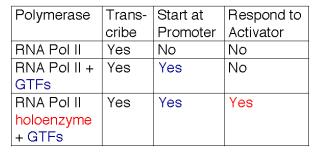
Fig.3.1.17
If the holoenzyme is the primary enzyme involved in transcription initiation in eukaryotic cells, then the progressive assembly pathway observed in vitro (see section d above) may be of little relevance in vivo. Perhaps the holoenzyme will bind to promoters simply marked by binding of TBP (or TFIID) to the TATA box, in contrast to the progressive assembly model that has a more extensive, ordered assembly mechanism. In both models, TBP or TFIID binding is the initial step in assembly of the preinitiation complex. However, at this point one cannot rule out the possibility that the holoenzyme is used at some promoters, and progressive assembly occurs at others.
7. Targets for the activator proteins
The targets for transcriptional activator proteins may be some component of the initiation complex. One line of investigation is pointing to the TAFs in TFIID as well as TFIIB as targets for the activators. Thus the activators may facilitate the ordered assembly of the intiation complex by recruiting GTFs. However, the holoenzyme contains the "mediator" or SRB complex that can mediate response to activators. Thus the activators may serve to recruit the holoenzyme to the promoter. Further studies are required to establish whether one or the other is correct, or if these are separate paths to activation.
F. General transcription factors for eukaryotic RNA polymerases I and III
1. General transcription factors for RNA polymerase I
a. Core promoter covers the start site of transcription, plus an upstream control element located about 70 bp further 5'.
b. The factor UBF1 binds to a G+C rich sequence in both the upstream control element and in the core promoter.
c. A multisubunit complex called SL1 binds to the UBF1‑DNA complex, again at both the upstream and core elements.
d. One of the subuntis of SL1 is TBP ‑ the TATA‑binding protein from TFIID!
e. RNA polymerase I then binds to this complex of DNA+UBF1+SL1 to initiate transcription at the correct nucleotide and the elongate to make pre‑rRNA.
2. General transcription factors for RNA Pol III
a. Internal control sequences are characteristic of genes transcribed by RNA Pol III (see below).
b. TFIIIA: binds to the internal control region of genes that encode 5S RNA (type 1 internal promoter)
c. TFIIIC: binds to internal control regions of genes for 5S RNA (alongside TFIIIA) and for tRNAs (type 2 internal promoters)
d. TFIIIB: The binding of TFIIIC directs TFIIIB to bind to sequences (-40 to +11) that overlap the start site for transcription. One subunit of TFIIIB is TBP, even though no TATA box is required for transcription. TFIIIA and TFIIIC can now be removed without affecting the ability of RNA polymerase III to initiate transcription. Thus TFIIIA and TFIIIC are assembly factors, and TFIIIB is the initiation factor.
Figure 3.1.18.
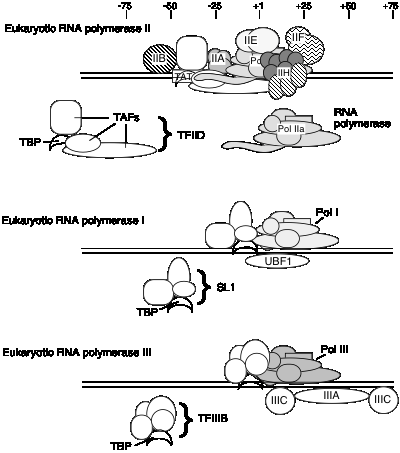
e. RNA polymerase III binds to the complex of TFIIIB+DNA to accurately and efficiently initiated transcription.
3 Transcription factor used by all 3 RNA Pol'ases: TBP
TBP seems to play a common role in directing RNA polymerase (I, II and III) to initiate at the correct place. The multisubunit factors that contain TBP (TFIID, SL1 and TFIIIB) may serve as positioning factors for their respective polymerases.
Questions for Chapter 10. Transcription: RNA polymerases
10.1 What is the role of the sigma factor in transcription, and how does it accomplish this?
10.2 Specific binding of E. coli RNA polymerase to a promoter:
1) completely envelopes the DNA duplex (both sides).
2) requires sigma factor to be part of the holoenzyme.
3) is enhanced by methylation of purine bases.
4) results in a temperature-dependent unwinding of about 10 base pairs.
Which statements are correct?
10.3 (POB) RNA polymerase. How long would it take for the E. coli RNA polymerase to synthesize the primary transcript for E. coli rRNAs (6500 bases), given that the rate of RNA chain growth is 50 nucleotides per second?
10.4 What is the maximum rate of initiation at a promoter, assuming that the diameter of RNA polymerase is about 204 Angstroms and the rate of RNA chain growth is 50 nucleotides per second?
10.5 Although three different eukaryotic RNA polymerases are used to transcribe nuclear genes, the enzymes and their promoters show several features in common. Are the following statements about common features of the polymerases and their mechanisms of initiation true or false?
a) All three purified polymerases need additional transcription factors for accurate initiation at promoter sequences.
b) All three polymerases catalyze the addition of a nucleotide "cap" to the 5' end of the RNA.
c) For all three polymerases, the TATA‑binding protein is a subunit of a transcription factor required for initiation (not necessarily the same factor for each polymerase).
d) All three polymerases are composed of multiple subunits.
10.6 What is common and what
is distinctive to the reactions catalyzed by DNA polymerase, RNA polymerase,
reverse transcriptase, and telomerase?