B
M B 400, Part Three
Gene
Expression and Protein Synthesis
Section
V = Chapter 14
TRANSLATION
A reminder: mRNA encodes the polypeptide with each amino acid designated by a string of three nucleotides. tRNAs serve as the adaptors to translate from the language of nucleic acids to that of proteins. Ribosomes are the factories for protein synthesis.
Figure 3.5.1.
|
|
A. tRNAs
1. The transfer RNAs, or tRNAs serve as adaptors to align the appropriate amino acids on the mRNA templates.
2. Primary structure of tRNAs
a. tRNAs are short, being only 73 to 93 nts long.
b. All tRNAs have the trinucleotide CCA at the 3' end.
(1) The amino acid is attached to the terminal A of the CCA.
(2) In most prokaryotic tRNA genes, the CCA is encoded at the 3' end of the gene. No known eukaryotic tRNA gene encodes the CCA, but rather it is added posttranscriptionally by the enzyme tRNA nucleotidyl transferase.
c. tRNAs have a large number of modified bases.
Over 50 different post‑transcriptional covalent modifications are known in tRNAs, such as dihydrouridine (D), in which the double bond between C4 and C5 is reduced, or pseudouridine (y), in which C5 is replaced with a N, providing another endocyclic amino group. The modified bases are especially prevalent in the loops.
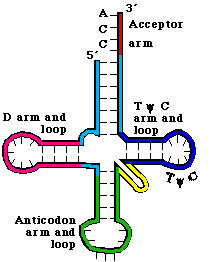
Figure
3.5.2. Secondary structure of tRNA.
3. The secondary structure of tRNA is a cloverleaf
a. tRNAs have 4 arms with 3 loops (see Figure 3. 5.2. for yeast phenylalanine tRNA)
b. The amino acid acceptor arm is formed by complementary base‑pairing between the intial 7 nts of tRNA and a short segment near the 3' end. Again, the amino acid will be added to the terminal A.
c. The D arm ends in the D loop. It contains several dihydrouridines, which are abbreviated "D".
d. The anticodon arm ends in anticodon loop. The anticodon is located in the center of the loop. It will align 3' to 5' with the mRNA (reading 5' to 3').
e. The variable loop varies in size in different tRNAs. The difference in size between the 73 nt versus 93 nt tRNAs is found in the variable loop.
f. The TyC arm is named for this highly conserved motif found in the loop.
4. The tertiary structure of tRNA
is a "fat L".
See Fig 3.5.3.
a. Some nucleotides in the D loop form base pairs with some nucleotides in the TyC loop. These and other interactions bring the cloverleaf (secondary structure) into an inverted L shape, with the "additional" base pairs found mainly at the junction of the inverted L.
b. In the 3‑D structure, two RNA double helices are at right angles. One of the double helices is the TyC stem in line with the amino acid acceptor stem. The other double helix has the D stem in line with the anticodon stem.
c. The result is that the two "business ends" of the tRNA are widely separated in space, at the two extremes of the tRNA. That is, the amino acid acceptor site is maximally separated from the anticodon (Figs. 3.5.3).
d. The rest of the molecule is a complex surface that must be recognized accurately by aminoacyl‑tRNA synthetases.
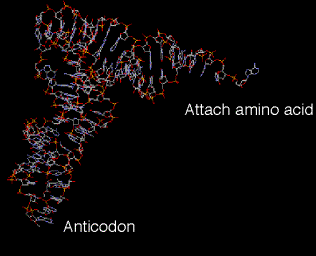
Fig. 3.5.3.
3-D structure of tRNA
A chime tutorial on tRNA structure is available from Dr. William McClureÕs website at Carnegie-Mellon University:
http://info.bio.cmu.edu/Courses/BiochemMols/tRNA_Tour/tRNA_Tour.html
B. Attachment of amino acids to tRNA
1. Aminoacyl‑tRNA synthetases
a. Approximately 20 enzymes, one per amino acid.
b. Must recognize several cognate tRNAs, i.e. that accept the same amino acid but recognize a different codon in the mRNA (a consequence of the degeneracy in the genetic code).
c. Must not recognize the incorrect tRNA ‑ i.e. these enzymes require precise discrimination among tRNAs.
d. Two different classes of aminoacyl‑tRNA synthetases
The two classes of enzymes are distinguished by the structure of their tRNA‑binding regions. The different classes of enzyme approach and bind to different faces of the tRNA, but both must recognize the ends as well as any distinguishing features of the their cognate tRNAs.
Each class has about ten synthetases (for ten amino acids).
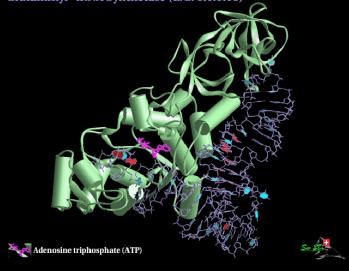
Fig. 3.5.4. 3-D structure of Glutaminyl-tRNA synthetase
The two classes of enzymes do not resemble each other much at all, in either sequence or 3-D structure, leading to the suggestion that they have evolved separately. If so, this would imply that an early form of life may have evolved using the ten amino acids handled by one class (or the other) of synthetase.
2. Mechanism
a. Aminoacyl-tRNA synthetase catalyzes a 2 step reaction. (Fig. 3.5.5)
First the amino acid is activated by adenylylation, i.e. a mixed anhydride intermediate is formed between the COO- of the amino acid and the a‑phosphoryl of ATP, with the liberation of pyrophosphate. The intermediate (activated amino acid) is an aminoacyl‑AMP..
In the second step, the amino acid is transferred to the 3' (or 2') OH of the ribose of the terminal A of tRNA, with liberation of AMP.
b. The product aminoacyl‑tRNA retains a high energy bond in an ester linkage.
(a) The equilibrium constant is about 1 for each of the two reactions, so the high energy of the bond initially between the a and b phosphoryls of ATP is essentially still present in the ester between the amino acid and the ribose of tRNA.
(b) The high energy bond in aminoacyl‑tRNA provides a driving force for protein synthesis.
c. Hydrolysis of pyrophosphate (abbreviated PPi) to two phosphates provides the free energy to drive synthesis of the aminoacyl-tRNA.
Thus one can consider that the equivalent of 2 ATPs (i.e. two high energy bonds) are used to form aminoacyl-tRNA, but one of the high energy bonds is retained in the product.
ATP ‑> AMP + PPi
PPi ‑> 2 Pi
In both instances, the cognate tRNA must be bound before proofreading can occur.

Figure 3.5.5.
3. Precise discrimination by AA‑tRNA synthetases
a. These enzymes must recognize the correct tRNA and the correct amino acid at the initial binding steps.
b. Proofreading is the removal of the incorrect amino acid (or tRNA) after binding, and often after part of the enzymatic reaction has occurred.
This can occur at either of the two reactions ‑ some synthetases will cleave an incorrect aminoacyl‑adenylate intermediate, and others will add the incorrect amino acid to the tRNA before recognizing the mistake and cleaving off the incorrect amino acid.
C.
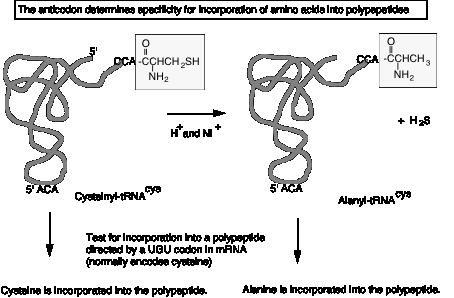
Anticodon determines specificity
The anticodon determines specificity for incorporation into a polypeptide during translation, not the amino acid. This was shown in the following experiment.
a. Cys‑tRNAcys can be converted to Ala‑tRNAcys by reductive desulfuration (H+ and Raney nickel), releasing H2S.
b. The resultant Ala‑tRNAcys retains the ACA anticodon to match a UGU codon in mRNA. When tested in cell‑free translation, it causes alanine to be incorporated instead of cysteine. (Fig. 3.4.4.)
c. Thus the amino acid on the tRNA did not direct its incorporation into the growing polypeptide chain, the anticodon did.
Figure
3.5.6.
D. Special tRNA for intiation of translation
1. Although Met has a single codon, two different tRNAs with different functions recognize the AUG codon.
(1) tRNAfmet (often abbreviated tRNAf) is used for initiation or translation in bacteria. A comparable initiator tRNA, called tRNAi , is used in eukaryotes.
(2) tRNAmmet is used for elongation.
Figure 3.5.5.
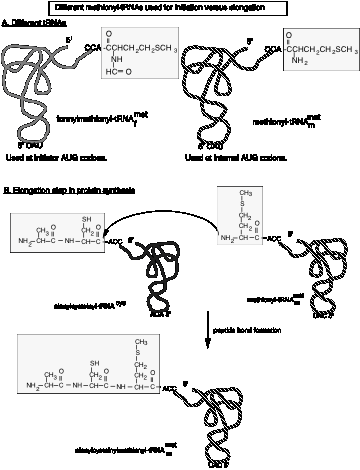
2. In bacteria, a formyl group is added to the amino group on the charged Met‑tRNAf , using 10‑formyl‑tetrahydrofolate as the formyl donor. This prevents its use in elongation.
3. In bacteria, only formylmethionyl‑tRNAf can bind to the partial P site on the small ribosomal subunit (see below) to initiate translation at AUG, or GUG (less frequently) or UUG (rarely). In all three cases, the protein starts with formylmethionine. The formyl group is removed after the first several amino acids have been incorporated, and in about half the cases, the methionine is also removed.
4. Note that the meaning of AUG and GUG is dependent on the context. AUG or GUG at the initiation site encodes formyl‑Met, but when internal to the mRNA, they encode Met or Val, respectively.
5. tRNAf has a different structure from tRNAm, and these differences determine their use either in initiation or elongation.
6. In eukaryotes, Met‑tRNAi is used for initiation. Although it is not formylated, the basic process is similar to that in prokaryotes.
E. Ribosomes
1. Role of ribosomes
a. Ribosomes are the molecular machines that catalyze peptide bond formation between a growing polypeptide and an incoming aminoacyl‑tRNA. The ribosomes insures that the amino acids are added in the order specified by the mRNA.
b. Ribosomes associate reversibly with the mRNA.
The two subunits of the ribosome form a complex around the mRNA to translate, and then dissociate after translation is completed.
2. Size and Composition of large and small subunits (see Fig.3.5.6.).
a. Ribosomes ("ribonucleic acid" "bodies") are large complexes of RNA and protein, with a roughly 60:40 ratio between RNA and protein. There are two subunits. Similar components are found in both eukaryotes and prokaryotes, although their sizes differ.
b. Each subunit has one major RNA (in bacteria, 23S rRNA for the large subunit and 16S for the small subunit) and many proteins (31 and 21, respectively, for bacterial large and small subunits). The large subunit also has a small rRNAs about 120 nucleotides in size (5S RNA). Eukaryotic large ribosomal subunits have an additional small RNA (5.8S) that corresponds to the sequence of the 5' end of bacterial 23S rRNA.
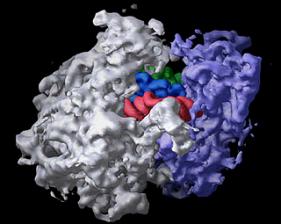
The bacterial ribosome is composed of three different RNA molecules and more than 50 different proteins arranged in two major subunits, which join together to form the complete ribosome. During protein synthesis, the ribosome binds transfer RNA molecules in three different sites. In this image of the ribosome with transfer RNAs in all three binding sites, the large subunit is gray, the small subunit is violet, and the three transfer RNAs are green, blue, and red. Image is from the Center for Molecular Biology of RNA, http://currents.ucsc.edu/99-00/09-27/ribosome.art.html
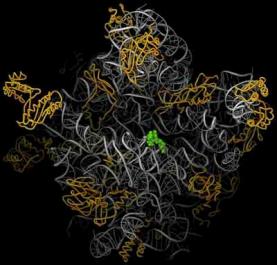
Figure 3.5.8. Images of ribosomes based on 3-D structure determination. The top view is from the Noller lab at UCSC, the bottom is from the Steitz lab and collaborators at Yale. The bottom view shows the RNA in silver ribbons and protein as gold coils. A green tRNA is at the peptidyl transferase site. Image from http://www.npaci.edu/features/01/05/05_03_01.html
c. The rRNAs and subunits were initially characterized by their sedimentation velocity, and hence are referred to by their sedimentation value in Svedberg units, or S. Larger macromolecules and complexes sediment faster and have a higher S value. However, other factors play a role in sedimentation rate (such as shape) and the S values for a complex is not the sum of the S values of individual components.
3. Shape
a. The small subunit is fairly elongated and binds mRNA.
b. The large subunit is more spherical and covers the small subunit.
c. The mRNA may thread between the 2 subunits or it may lie outside the ribosome.
4. P (peptidyl‑tRNA) and A (aminoacyl‑tRNA) and E (exit) sites
A tRNA interacts with the ribosome at three major sites as it brings in an amino acid, has the growing polypeptide chain attached to that amino acid, and then finally leaves the ribosome after donating its amino acid.
a. A site (or entry site): aminoacyl‑ tRNA binds
b. P site (or donor site): peptidyl‑tRNA binds, i.e. the nascent polypeptide chain linked to the last tRNA to occupy the A site (see below).
c. E site: exit of deacylated tRNA after peptide bond formation.
d. Flow of tRNA through the ribsoome is from the A site to P site, then exit via the E site.
e. The next point will become clearer after we discuss the elongation cycle. The molecule attached to the 3' end of the tRNA is different at each site.
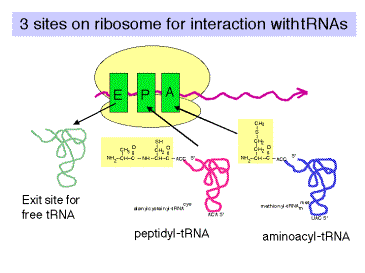
Fig. 3.5.9.
F. The polarity of translation is from the amino (N) terminus to the caboxy (C) terminus.
This was demonstrated in a classic experiment by Dintzis.
1. Actively translating proteins were labeled with radioactive amino acids for a brief time (short relative to the time required to complete synthesis).
2. Completed polypeptides were collected, digested with trypsin, and the amount of radioactivity in tryptic fragments was determined.
3. Tryptic fragments from the C‑terminal end of the polypeptide had radioactivity at the earliest times of labeling.
4. As the period of labeling was increased (longer pulse), tryptic fragments closer to the N terminus were labeled.
5. This shows that the direction of polypeptide growth is from the N teminus to the C terminus, i.e. translation begins at the N terminal amino acid. This corresponds to mRNA chain growth in a 5' to 3' direction.
6. Note that this experimental protocol is also used to map origins of replication, as we covered in Part Two of the course.
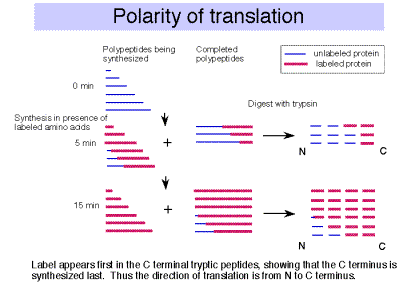
Fig.
3.5.10.
G. Initiation of translation
1. mRNA binds to small ribosomal subunit (not the whole 50S ribosome) in such a way that the initiator AUG is positioned in the precursor to the P site, i.e. ready for the f-met-tRNAfmet to recognize it.
a. The alignment of the initiator AUG in the mRNA with the appropriate place on the ribosomal subunit involves base pairing between the 3' end of 16S rRNA and a sequence that precedes the initiator AUG in mRNA. When this portion of 16S rRNA in the 23S subunit is removed by cleavage with colicin (an antibiotic), the 23S subunit loses the ability to initiate translation.
Figure 3.5.11.
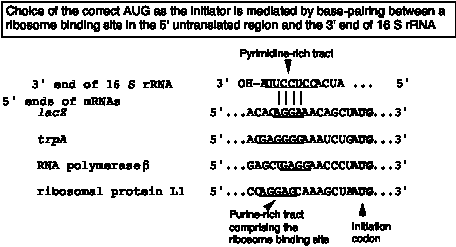
b. The ribosome binding site is in the 5' untranslated region, just before the initiator AUG. It is also called a Shine‑Dalgarno sequence (named for the discoverers of the sequence).
It is a purine‑rich sequence, e.g. 5' AGGAG, that will pair with the pyrimidine‑rich 3' end of 16S rRNA (5' CCUCCUUA‑OH 3')
c. This base pairing insures the choice of the correct AUG as initiation codon, as opposed to an internal AUG.
2. Roles of initiation factors and other factors
a. Translation factors are used at only one step of the process and are not permanent subunits of the ribosome. They cycle on and off the ribosomes as they do their function. They are (frequently) present in smaller amounts than the ribosomal subunits.
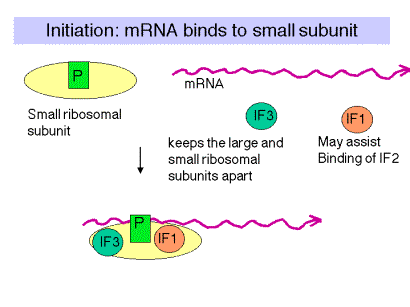
Figure 3.5.12.
b. IF3 = Initiation Factor 3
(1) An antiassociation factor; prevents association between the large and small ribosomal subunits.
(2) It also must be associated with the small subunit for it to form an initiation complex, i.e. for the small subunit to correctly bind mRNA and fmet-tRNAf.
(3) It dissociates prior to binding of the large subunit.
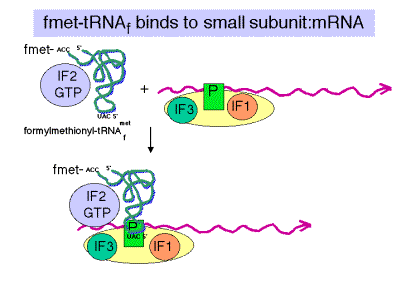
Fig. 3.5.13.
c. IF2
(1) Brings fmet‑tRNAf to the partial P site on the small subunit.
(2) At least in eukaryotes, it does this in a ternary complex with IF2, fmet‑tRNAf and GTP. In bacteria, the GTP may bind the initiation complex separately. [In some texts, such as MBOG, p. 412, the GTP-IF2 complex binds to the 30S subunit separately from fmet-tRNAf. How would you test the differences in these two models?]
(3) IF2 activates a GTPase activity in the small subunit. The resulting change in conformation may allow the large subunit to bind.
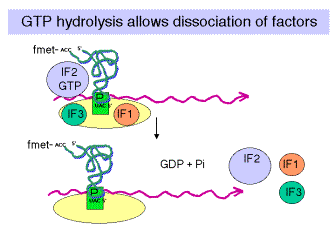
Fig. 3.5.14.
d. GTP
Hydrolysis, stimulated by IF2, promotes dissocation of IF2, IF1 and IF3 from the initiation complex and association of the 50S subunit.
e. IF1: role is unknown; perhaps it is an assembly factor that assists in the binding of IF2.
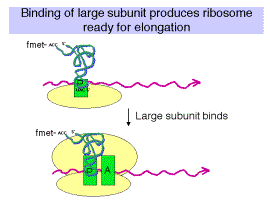
Fig. 3.5.15.
3. Binding of 50S (large) subunit to initiation complex gives a complete ribosome ready for the elongation phase of translation. Note that f-met-tRNAfmet is positioned at the P site. It has recognized the initiator AUG in the mRNA.
4. Identification of initiator AUG in eukaryotes
a. Bases around AUG influence efficiency of initiation.
(1) The most important effects are from a purine 3 nt before AUG and a G after it. The preferred context is RNNAUGG.
(2) The consensus sequence for a large number of mRNAs is GCCRCCAUGG, but these other nucleotides have little effect in mutagenesis experiments.
a. Modified scanner model
(1) The mRNA is "prepared" for binding to the ribosome by the action of eukaryotic initiation factor 4, abbreviated eIF4 (Fig. 3.5.16). eIF4 is a multisubunit factor; it includes a cap‑binding protein, eIF4F, that recognizes the 5' cap structure. It also includes proteins eIF4A and eIF4B. These are RNA helicases, which unwind secondary structures in the 5' untranslated region of the mRNA at the expense of ATP hydrolysis.
The mRNA then binds to the small ribosomal subunit. The met-tRNAi has already been brought to the small ribosomal subunit by eIF2, in a complex with GTP.
eIF3 keeps the small ribosomal subunit apart from the large subunit during the process of binding the mRNA.
(2) The small subunit, with associated factors, scans along the mRNA until it reaches (usually) the first AUG. Factors eIF1 and eIF1A help move the preinitiation complex to the AUG start.
Fig. 3.5.16.
H. The elongation cycle during translation
1. Binding of aminoacyl‑tRNA to the A site
Recent review: Weijland, A. and A. Parmeggiani (1994) TIBS 19:188-193.
Schroeder, R. (1994) Nature 370:597.
a. Elongation factor EF‑Tu
(1) The ternary complex of aminoacyl‑tRNA, EF‑Tu, and GTP brings the aminoacyl‑tRNA to the A site on the 70S ribosome (fig. 3.5.17).
(2) After the aminoacyl‑tRNA is deposited at the A site of the ribosome, the GTP is cleaved to GDP + Pi. The binary complex of EF‑Tu and GDP dissociates from the ribosome.
(3) This is one of many examples of guanine‑nucleotide‑binding proteins that are active when GTP is bound and inactive when GDP is bound.
The general model is that the GTP-bound state of EF-Tu adopts a conformation with a high affinity for aminoacyl-tRNA. The conformation (shape, charge density, etc.) of the resulting ternary complex (containing EF-Tu,GTP, and aminoacyl-tRNA) then allows it to bind to the A site of the ribsosome. Hydrolysis of GTP to form GDP and inorganic phosphate causes the EF-Tu to adopt a different conformation. The aminoacyl-tRNA now has a lower affinity for EF-Tu in the GDP bound state, and presumably a higher affinity for the A site on the ribosome, so it stays on the ribosome when EF-Tu in the GDP bound state dissociates (both from aminoacyl-tRNA and from the ribosome).
Figure 3.5.17.
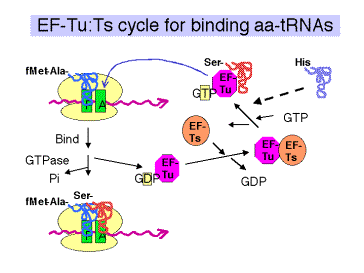
(4) EF‑Tu is one of the most abundant proteins in E. coli, at 70,000 copies per cell. This is almost equal to the number of aminoacyl‑tRNAs per cell, so most of the aminoacyl‑tRNAs are likely to be in the ternary complex when the concentration of GTP is sufficiently high.
b. GTP
(1) Required for binding aminoacyl‑tRNA.
(2) Hydrolysis promotes dissociation of the complex EF‑Tu plus GDP from the ribosome.
c. EF‑Ts
(1) Aids in the recycling of EF‑Tu by GDP‑GTP exchange.
(2) EF‑Ts binds to EF‑Tu complexed with GDP, causing dissociation of GDP. GTP can now bind to the EF‑Tu‑Ts complex, causing EF‑Ts to dissociate and leaving EF‑Tu complexed with GTP. This latter binary complex is ready to bind another aminoacyl‑tRNA.
d. The antibiotic kirromycin prevents release of EF‑Tu‑GDP, thereby blocking elongation. This demonstrates that one step must be completed before the next can take place, and illustrates the importance of the EF‑Tu‑GTP/GDP cycle.
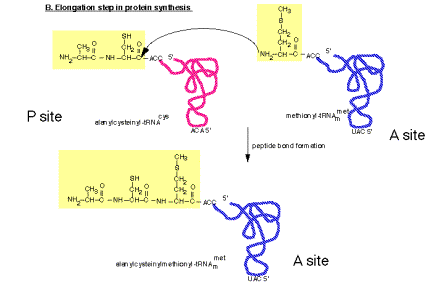
2. Peptidyl transferase on the large ribosomal subunit
a. The peptidyl transferase reaction occurs via nucleophilic displacement. The amino group from aminoacyl‑tRNA (position n) attacks the "C‑terminal" carboxyl group of peptidyl‑tRNA (position n‑1 in the mRNA).
This results in cleavage of the high energy peptidyl‑tRNA ester linkage, thereby providing the free energy to drive the reaction.
The resulting products of the reaction are deacylated tRNA at the P site and peptidyl‑tRNA at the A site.
Figure 3.5.18. Peptidyl transferase reaction
b. Role of rRNA in catalysis
It is likely that rRNA provides the catalytic center for the peptidyl transferase activity, with perhaps some ribosomal proteins aiding in holding the rRNA in the correct conformation for catalysis. This conclusion is supported by several lines of investigation, some of which are listed below.
(1) No protein, singly or in combination with other proteins, has been shown to catalyze peptide bond formation.
(2) Specific regions of 16S rRNA (in the small subunit) interact with the anticodon regions of tRNA in both the A and P sites. In contrast, 23S rRNA in the large subunit interacts with the CCA terminus of peptidyl‑tRNA, thus placing it in the right location for peptidyl transferase.
(3) The antibiotics erythromycin and chloramphenicol block peptidyl transferase. Some mutations that confer resistance to them map to the 23S rRNA sequence (others map to some 50S ribosomal proteins).
(4) A preparation consisting of 23S rRNA and some remnants of large subunit proteins retains peptidyl transferase activity. For more information, see Noller et al. (1992) Unusual resistance of peptidyl transferase to protein extraction procedures. Science 256: 1416-1419.
(5) Ribozyme RNAs can be selected that catalyze peptide bond formation. In this experiment, the investigators started with a pool of 1.3 « 1015 different RNAs of 72 nucleotides, flanked by constant regions. They let this large population of RNAs catalyze a peptide bond formation that adds a biotinyl-labeled amino acid (in a chemical mimic of a P site) to an amino acid connected to the RNA (in a chemical mimic of an A site). The RNAs that successfully catalyzed the reaction were extremely rare, but were now covalently attached to a biotin label. Thus they could be selected from the population by binding to streptavidin. PCR was used to amplify the successful RNAs, and the procedure repeated 19 times. At this point, the investigators characterized 9 RNAs that catalyzed the reaction. They found that these RNAs increased the reaction rate by a factor of 106 over the uncatalyzed reaction.
(6) The three-dimensional structure of the ribosome shows that the active site is comprised of RNA. The structure of a ribosome crystallized with an active site directed inhibitor has been determined, as well as the structure without the inhibitor. This allowed researchers to see precisely where the peptidyl transferase active site is within the structure. Only RNA is seen around this site. The nearest protein is 20 Angstroms away, too far to participate in catalysis.
3. Translocation
a. The translocation step moves the ribosome another 3 nucleotides downstream (one codon) and moves peptidyl‑tRNA to the P site (position n), deacylated tRNA exits through the E site, and the A site (position n+1) is vacant for another round of elongation.
b. Elongation Factor G = EF‑G
(1) This is another very abundant protein, with about 20,000 copies per cell, which is equivalent to the number of ribosomes.
(2) EF‑G‑GTP binds to the ribosome to aid translocation, and is released upon GTP hydrolysis (GTPase is from some ribosomal component).
(3) Recent structural studies (from A. Dahlberg and colleagues) show that EF-G in the GTP-bound state has a shape similar to that of the ternary complex of EF-Tu, GTP and aminoacyl-tRNA. Like the latter ternary complex, EF-G in the GTP-bound state also has a high affinity for the A site on the ribosome. This may help drive the movement of the peptidyl-tRNA from the A site to the P site, replacing it with EF-G (GTP) in the A site.
c. Hydrolysis of GTP is required for dissociation of EF‑G after translocation. The GTPase is part of the ribosome, not EF-G.
Fig.
3.5.20.
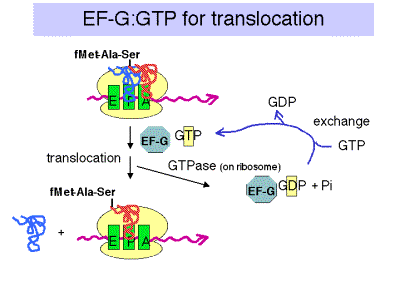
d. Action of fusidic acid revealed the need for release of EF‑G‑GDP.
In the presence of fusidic acid, EF‑G‑GTP binds the ribosome, GTP is hydrolyzed, and the ribosome moves three nucleotides. But the ribosome‑EF‑G‑GDP complex is stabilized by this compound, and translation is halted.
e. Ribosomes cannot bind EF‑Tu and EF‑G simultaneously.
EF‑Tu must finish its action before EF‑G can act, and EF‑G must complete its cycle before EF‑Tu can act again to bring in another aminoacyl‑tRNA.
f. Effect of diptheria toxin
(1) The eukaryotic analog to EF‑G is eEF2, which is also a translocase dependent on GTP hydrolysis. It is also is blocked by fusidic acid.
(2) Diptheria toxin will catalyze the addition of ADP‑ribose (from substrate NAD+) to eEF2, thereby inactivating it. The target for ADP‑ribosylation is modified histidine found in eEF2 from many species.
4. Elongation rate
a. Bacteria growing at 37o add about 15 amino acids to a growing chain each second.
b. In eukaryotes, the elongation rate is much slower, about 2 amino acids added per sec.
I. Termination
1. The three termination codons are:
UAG = amber
UAA = ochre; most common for bacterial genes
UGA = opal
2. Releasing factors (RF) are proteins that promote termination of translation and release of the mRNA from ribosomes at those termination codons.
a. Bacteria have two releasing factors
RF1 recognizes UAG and UAA
RF2 recognizes UGA and UAA
Figure 3.5.21.
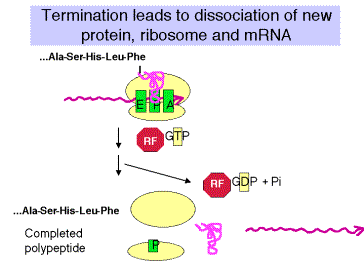
There are about 600 molecules of releasing factors per bacterial cell, or about 1 per 50 ribosomes. The releasing factors act when a termination codon is present at the ribosomal A site and peptidyl-tRNA is at the P site.
The
releasing factors may mimic the aminoacyl-tRNA in shape, but promote hydrolysis
of peptidyl-tRNA rather than transfer to a new aminoacyl-tRNA. Hence one
mechanism for the action of the releasing factors is to cause the ribosome to
use H20 as the nucleophile attacking the ester linkage of
peptidyl-tRNA, rather than the a-amino
group of the aminoacyl-tRNa acting as a nucleophile.
b. Eukaryotes:
In eukaryotes, a single releasing factor (eRF) has been characterized. This protein requires GTP to bind. Hydrolysis of GTP probably promotes dissociation of eRF from ribosomes.
3. GTP is utilized by eukaryotic releasing factors.
The theme of using accessory proteins that cycle on and off the ribosome in GTP-bound and GDP-bound states, respectively, is seen in initiation, at two steps in elongation, and now in termination. Curiously, it appears that the releasing factors in E. coli do not require GTP for their action. It is not clear that this represents a fundamental difference, since it is possible that the role of GTP hydrolysis has been adopted by some other ribosomal component. Indeed, the overall mechanism of translation has been highly conserved in all major groups of organisms (eubacteria, archaea, and eukaryotes).
4. Termination results in dissociation of the entire translation complex. This leads to three different releasing events:
a. Release of newly synthesized, completed polypeptide from the tRNA, which requires hydrolysis of the ester link between the nascent polypeptide and the tRNA.
b. Release of mRNA.
c. Release of the ribosome. When free, the ribosome is in equilibrium with the dissociated large and small subunits.
J. Mutant tRNAs can act as suppressors
1. Definition of suppressors
a. Mutations at a second site that can overcome a missense or nonsense mutation at an original site are suppressors. If the original mutation is reversed to wild type, this is called genotypic reversion, or a back-mutation. Both suppression and reversion will produce a wild type phenotype (at least a partial one) from an original mutation ‑ the distinction is whether the original mutation is changed (genotypic reversion), or whether a second site is altered (suppression).
b. The suppressor can be either intragenic or extragenic.
c. These second sites that can mutate to suppress an original mutation usually encode a cellular component that interacts with the component encoded by the originally mutated locus. Isolation of suppressors can be used to piece together pathways or complex cellular structures.
2. Nonsense suppressors
a. Nonsense suppressors are often mutant tRNAs that still accept an amino acid but whose anticodon has been altered to match a termination codon.
b. E.g. supE encodes a mutant tRNAgln
Figure 3.5.21.
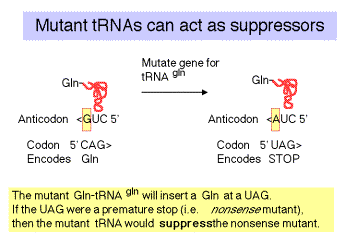
c. The "down side" to nonsense suppression is that the suppressor tRNA can act at any amber codon. Therefore it competes with the releasing factors in recognizing the normal termination codons. When the suppressor tRNA is used instead of releasing factors, translation proceeds further down the mRNA than it is supposed to, leading to production of aberrant proteins. Suppressor strains of E. coli can be pretty sick (i.e. they don't grow as well as wild type strains).
d. Two other amber suppressors are encoded by the supD gene, which encodes a tRNA that will insert Ser at a UAG, and supF, which will insert Tyr.
3. Missense suppressors
These are mutant tRNAs that lead to the insertion of an amino acid that is compatible with the wild type amino acid (altered by the original mutation).
4. Frameshift suppressors
These are mutant tRNAs whose anticodon has been expanded (or contracted?) to match the length‑altering mutation in the mRNA.
E.g. Consider an original mutation 5'GGG ‑> 5'GGGG (insert a G).
A frameshift suppressor would also have an additional C in the anticodon.
wt tRNA anticodon 3'CCC ‑‑> suppressor tRNA 3'CCCC.
Questions on Chapter 14. Translation
14.1 (POB) Methionine Has Only One Codon.
Methionine is one of the two amino acids having only one codon. Yet the single codon for methionine can specify both the initiating residue and interior Met residues of polypeptides synthesized by E. coli. Explain exactly how this is possible.
14.2 Are the following statements concerning aminoacyl‑tRNA synthetase true or false?
a) Two distinct classes of the enzymes have been defined that are not very related to each other.
b) The enzymes scan previously‑synthesized aminoacyl‑tRNAs and cleave off any amino acids that are linked to the incorrect tRNA.
c) Proofreading can occur at the formation of either the aminoacyl‑adenylate intermediate (in some synthetases) or at the aminoacyl‑tRNA (in other synthetases) to insure that the correct amino acid is attached to a given tRNA.
d) The product of the reaction has a high‑energy ester bond between the carboxyl of an amino acid and a hydroxyl on the terminal ribose of the tRNA.
14.3 A preparation of ribosomes in the process of synthesizing the polypeptide insulin was incubated in the presence of all 20 radiolabeled amino acids, tRNA's, aminoacyl-tRNA synthetases and other components required for protein synthesis. All the amino acids have the same specific radioactivity (counts per minute per nanomole of amino acid). It takes ten minutes to synthesize a complete insulin chain (from initiation to termination) in this system. After incubation for 1 minute, the completed insulin chains were cleaved with trypsin and the radioactivity of the fragments determined.
a) Which tryptic fragment has the highest specific activity?
b) In the same system described above, the insulin polypeptide chains still attached to the ribosomes after ten minutes were isolated, cleaved with trypsin, and the specific activity of each tryptic peptide determined. Which peptide has the highest specific activity?
14.4 Which component of the protein synthesis machinery of E. coli carries out the function listed for each statement.
a) Translocation of the peptidyl-tRNA from the A site to the P site of the ribosome.
b) Binding of f-Met-tRNA to the mRNA on the small ribosomal subunit.
c) Recognition of the termination codons UAG and UAA.
d) Holds the initiator AUG in register for formation of the initiation complex (via base pairing).
14.5 a) In the initiation of translation in E. coli, which ribosomal subunit does the mRNA initially bind to?
b) What nucleotide sequences in the mRNA are required to direct the mRNA to the initial binding site on the ribosome?
c) What other factors are required to form an initiation complex?
14.6 What steps in the activation of amino acids and elongation of a polypeptide chain require hydrolysis of high energy phosphate bonds? What enzymes catalyze these steps or which protein factors are required?
14.7 (POB) Maintaining the Fidelity of Protein Synthesis
The chemical mechanisms used to avoid errors in protein synthesis are different from those used during DNA replication. DNA polymerases utilize a 3' ¨ 5' exonuclease proofreading activity to remove mispaired nucleotides incorrectly inserted into a growing DNA strand. There is no analogous proofreading function on ribosomes; and, in fact, the identity of amino acids attached to incoming tRNAs and added to the growing polypeptide is never checked. A proofreading step that hydrolyzed the last peptide bond formed when an incorrect amino acid was inserted into a growing polypeptide (analogous to the proofreading step of DNA polymerases) would actually be chemically impractical. Why? (Hint: Consider how the link between the growing polypeptide and the mRNA is maintained during the elongation phase of protein synthesis.)
14.8 (POB) Expressing a Cloned Gene.
You have isolated a plant gene that encodes a protein in which you are interested. What are the sequences or sites that you will need to get this gene transcribed, translated, and regulated in E. coli.)?
14.9 The three codons AUU, AUC, and AUA encode isoleucine. They correspond to "hybrid" between a codon family (4 codons) and a codon pair (2 codons). The single codon AUG encodes methionine. Given the prevalence of codon pairs and families for other amino acids, what are hypotheses for how this situation for isoleucine and methionine could have evolved?
14.10 Use the following processes to answer parts a-c:
[1] synthesis of aminoacyl-tRNA from an amino acid and tRNA.
[2] binding of aminoacyl-tRNA to the ribosome for elongation.
[3] formation of the peptide bond between peptidyl-tRNA and aminoacyl-tRNA on the ribosome.
[4] translocation of peptidyl-tRNA from the A site to the P site on the ribosome.
[5] assembly of a spliceosome for removal of introns from nuclear pre-mRNA.
[6] removal of introns from nuclear pre-mRNA after assembly of a spliceosome.
[7] synthesis of a 5' cap on eukaryotic mRNA.
(a) Which of the above processes require ATP?
(b) Which of the above processes require GTP?
(c) For which of the above processes is
there evidence that RNA is used as a catalyst?
