CHAPTER 5
DNA REPLICATION I:
Enzymes and mechanism
A fundamental property of living organisms is their ability to reproduce. Bacteria and fungi can divide to produce daughter cells that are identical to the parental cells. Sexually reproducing organisms produce offspring that are similar to themselves. On a cellular level, this reproduction occurs by mitosis, the process by which a single parental cell divides to produce two identical daughter cells. In the germ line of sexually reproducing organisms, a parental cell with a diploid genome produces four germ cells with a haploid genome via a specialized process called meiosis. In both of these processes, the genetic material must be duplicated prior to cell division so that the daughter cells receive a full complement of the genetic information. Thus accurate and complete replication of the DNA is essential to the ability of a cell organism to reproduce.
In this chapter and the next, we will examine the process of replication. After describing the basic mechanism of DNA replication, we discuss the various techniques researchers have used to achieve a more complete understanding of replication. Indeed, a theme of this chapter is the combination of genetic and biochemical approaches that has allowed us to uncover the mechanism and physiology of DNA replication. In the remaining sections of the chapter, we focus on the enzymes that mediate DNA replication. In these descriptions, you will encounter several cases of structure suggesting a particular function. We will point out parallels and homologies between bacterial and eukaryotic replication components. This chapter covers the basic process and enzymology of DNA synthesis, and the next chapter will cover regulation of DNA replication.
Basic Mechanisms of Replication
DNA replication is semiconservative.
We begin our investigation by describing the basic model for how nucleotides are joined in a specific order during DNA replication. By the early 1950’s, it was clear that DNA was a linear string of deoxyribonucleotides. At that point, one could postulate three different ways to replicate the DNA of a cell. First, a cell might have a DNA-synthesizing "machine" which could be programmed to make a particular string of nucleotides for each chromosome. A second possibility is that the process of replication could break the parental DNA into pieces and use them to seed synthesis of new DNA.
A third model could be proposed from the DNA structure deduced by Watson and Crick. When they described the double-helical structure of DNA in a one-page article in Nature in 1953, they included this brief statement of a third model:
"It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material."
A subsequent paper elaborated on this mechanism. The complementarity between base pairs (A with T and G with C) not only holds the two strands of the double helix together, but the sequence of one strand is sufficient to determine the sequence of the other. Hence a third possibility for a mechanism of DNA replication was clear - one parental strand could serve as a template directing synthesis of a complementary strand in the daughter DNA molecules. This 1953 paper is of course most famous for its description of the double-helical structure of DNA held together by base complementarity, but it is also important because the proposed structure suggested a testable model for how a particular process occurs, in this case replication.
These three models make different predictions about the behavior of the two strands of the parental DNA during replication (Fig. 5.1). In the first, programmed machine model, the two strands of the parental DNA can remain together, because they are not needed to determine the sequence of the daughter strands. This model of replication is called conservative: the parental DNA molecules are the same in the progeny as in the parent cell. In the second model, the each strand of the daughter DNA molecules would be a combination of old and new DNA. This type of replication is referred to as random (or dispersive). The third model, in which one strand of the parental DNA serves as a template directing the order of nucleotides on the new DNA strand, is a semiconservative mode of replication, because half of each parent duplex (i.e. one strand) remains intact in the daughter molecules.
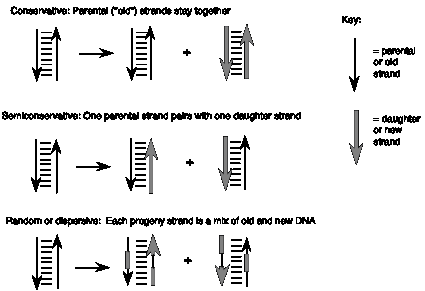
Figure 5.1. Possible models of replication of a duplex nucleic acid.
When they were graduate students at the California Institute of Technology, Matthew Meselson and Franklin Stahl realized that they could test these three models for replication by distinguishing experimentally between old and new strands of DNA. They labeled the old or parental DNA with nucleotides composed of a heavy isotope of nitrogen (15N) by growing E. coli cells for several generations in media containing [15N] NH4Cl. Ammonia is a precursor in the biosynthesis of the purine and pyrimidine bases, and hence this procedure labeled the nitrogen in the nucleotide bases in the DNA of the E. coli cells with 15N. The cells were then shifted to grow in media containing the highly abundant, light isotope of nitrogen, 14N, in the NH4Cl, so that newly synthesized DNA would have a "light" density. The labeled, heavy (old) DNA could be separated from the unlabeled, light (new) DNA on a CsCl density gradient, in which the DNA bands at the position on the gradient where the concentration of CsCl has a density equal to that of the macromolecule. At progressive times after the shift to growth in [14N] NH4Cl, samples of the cells were collected, then DNA was isolated from the cells and separated on a CsCl gradient.
A. B.
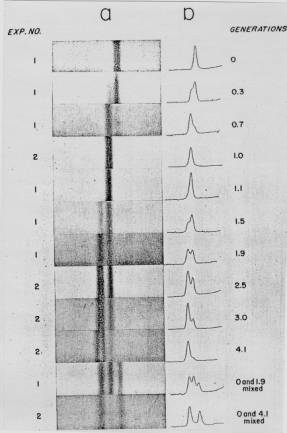
Figure 5.2. Results of the Meselson and Stahl experiment demonstrating semiconservative replication of DNA. A. The left panel (a) shows ultraviolet absorption photographs of DNA after equilibrium sedimentation in a CsCl gradient, as a function of the number of generations from the shift from media that labeled DNA with a high density (15N-labeled) to a medium in which the DNA is normal, or light density (14N-DNA). The density of the CsCl gradient increases to the right. The panel on the right (b) shows a trace of the amount of DNA along the gradient. The number of generations since the shift to the media with 14N substrates is shown at the far right. Mixing experiments at the bottom show the positions of uniformly light and heavy DNA (generations 0 and 4.1 mixed) and the mixture of those plus hybrid light and heavy DNA (generations 0 and 1.9 mixed). Parental DNA forms a band at the heavy density (15N-labeled), whereas after one generation in light (14N) media, all the DNA forms a band at a hybrid density (between heavy and light). Continued growth in light media leads to the synthesis of DNA that is only light density. B. The interpretation of the experimental results as demonstrating a semiconservative model of replication. Part A of this figure is Fig. 4 and Part B is Fig. 6 from M. Meselson and F. Stahl (1958) “The Replication of DNA in Escherichia coli” Proceedings of the National Academy of Sciences, USA 44:671-682.
The results fit the pattern expected for semiconservative replication (Fig. 5.2). To quote from Meselson and Stahl, “until one generation time has elapsed, half-labeled molecules accumulate, while fully labeled DNA is depleted. One generation time after the addition of 14N, these half-labeled or ‘hybrid’ molecules alone are observed. Subsequently, only half-labeled DNA and completely unlabeled DNA are found. When two generation times have elapsed after the addition of 14N, half-labeled and unlabeled DNA are present in equal amounts.” A conservative mode of replication is ruled out by the observation that all the DNA formed a band at a hybrid density after one generation in the [14N] NH4Cl-containing medium. However, it is consistent with either the semiconservative or random models. As expected for semiconservative replication, half of the DNA was at a hybrid density and half was at a light density after two generations in [14N] NH4Cl-containing medium. Further growth in the 14N medium resulted in an increase in the amount of DNA in the LL band.
Question 5.1: What data from this experiment rule out a random mode of replication?
These experiments demonstrated that each parental DNA strand is used as a template directing synthesis of a new strand during DNA replication. The synthesis of new DNA is directed by base complementarity. The enzymes that carry out replication are not programmed “machines” with an inherent specificity to synthesize a given sequence, but rather the template strand of DNA determines the order of nucleotides along the newly synthesized DNA strand (Fig. 5.3).
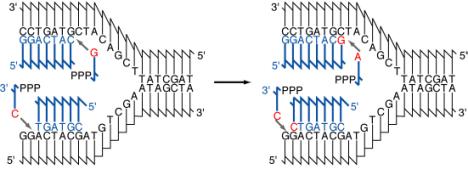
Fig. 5.3. Diagram of the addition of nucleotides in a new strand of DNA during semiconservative replication. The parental DNA strands are shown in black and the new DNA strands and deoxyribonucleoside triphosphates are in blue. The DNA strands are shown using the convention that vertical lines are the deoxyribose portion of each deoxyribonucleotide, and the connecting lines represent the phosphodiester linking the 3’ hydroxyl of one deoxyribonucleotide with the 5’ hydroxyl of the next. The part of the connecting line representing the 3’ end of the phosphodiester attached to the vertical (deoxyribose) line about 1/3 of the way along it, and the part of the connecting line representing the 5’ end of the phosphodiester is attached at the end of the vertical line. Bases are abbreviated by a single letter. The bases on the deoxyribonucleotides that are being added are in red. Two rounds of addition of nucleotides are shown. In this diagram, each strand of the parental DNA is serving as the template for synthesis of a new DNA strand. The chemistry of the synthesis reaction, the enzymes needed for separating the two parental strands, and other features of replication will be discussed later in the chapter.
The association of a parental DNA strand with a newly synthesized DNA strand observed in this important experimental result is consistent with the use of each parental DNA strand as a template to direct the replication machinery to place nucleotides in a particular order. Watson and Crick proposed that base complementarity would guide the replication machinery to insert an A opposite a T, a T opposite an A, a G opposite a C, and C opposite a G (Fig. 5.3). This was verified once the enzymes carrying out DNA synthesis were isolated, and the chemical composition of the products of replication was compared with that of the templates. These enzymes are discussed in detail later in the chapter, as will be the chemistry of the process of adding individual nucleotides to the growing DNA chain (a process called elongation). You may recall that these enzymes were also used to demonstrate the antiparallel arrangement of the DNA strands predicted by Watson and Crick (recall problem 2.5). With this understanding of how the sequence of nucleotides is specified, we can examine the types of DNA structures found during replication.
Specialized DNA structures are formed during the process of replication.
The process of semiconservative replication illustrated in Fig. 5.3 requires that the two strands of the parental DNA duplex separate, after which they serve as a template for new DNA synthesis. Indeed, this allows the same base-pairing rules and hydrogen-bonding patterns to direct the order of nucleotides on the new DNA strand and to hold the two strands of duplex DNA together. The region of replicating DNA at which the two strands of the parental DNA are separated and two new daughter DNA molecules are made, each with one parental strand and one newly synthesized strand, is called a replication fork. Once DNA synthesis has initiated, elongation of the growing new DNA strand proceeds via the apparent movement of one or two replication forks. The replication fork(s) are at one or both ends of a distinct replicative structure called a replication eye or bubble, which can be visualized experimentally (Fig. 5.4a). Examination of replicating DNA molecules in the electron microscope shows regions where a single DNA duplex separates into two duplexes (containing newly synthesized DNA) followed by a return to a single duplex. This has the appearance of an eye or a bubble, and hence the structure is named accordingly. The replication bubble can result from either bidirectional or unidirectional replication (Figure 5.4b). In bidirectional replication, two replication forks move in opposite directions from the origin, and hence each end of the bubble is a replication fork. In unidirectional replication, one replication fork moves in one direction from the origin. In this case, one end of a replication bubble is a replication fork and the other end is the origin of replication. If the chromosome is circular, the replication bubble makes a q structure. As replication proceeds, the emergent daughter molecules (composed of one old strand and one new strand of DNA) are the identical to each other, ever increasing size, whereas the unreplicated portion of the chromosomes becomes smaller and smaller.
A.
B.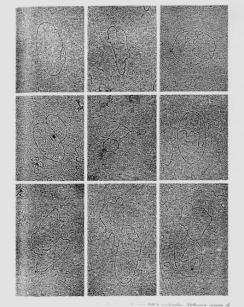
C. 
Figure 5.4. Replication bubbles. Panel A shows diagrams of the replication bubbles or “eyes” that form when the two parental template chains are separated and copied during replication. Replication bubbles in a circular DNA molecule resemble the Greek letter theta, or q. Panel B shows electron micrographs of replicating polyoma virus DNA. The viral DNA from polyoma is duplex, circular and relatively small (about 5000 bp), which facilitates resolution of the parts of the replicating molecules. Each molecule in this panel shows two branch points, which are replication forks for polyoma, and three branches. Two of the branches in each molecule are the same length; these are the newly replicated portions of the DNA. The pictures are arranged to show progressively more replication. This is a copy of plate I from B. Hirt (1969) “Replicating Molecules of Polyoma Virus DNA”, Journal of Molecular Biology 40:141-144. Panel C illustrates that a replication bubble can result from either unidirectional or bidirectional replication. The origin of replication is labeled ori.
One could imagine making a new daughter DNA molecule via semiconservative replication by completely separating the two strands of a DNA molecule, and then using each separated strand as a template to make two daughter molecules that were separate during the entire process of replication. However, the visualization of replication bubbles during replication shows that the daughter DNA molecules are still connected to the parental molecule, producing the characteristic "eye" form (Fig. 5.4). Hence the separation of the two strands is localized to the replication fork. Although we discuss replication in terms of moving replication forks, it is more likely that the forks are stationary at a complex replication site, and the DNA is moved through this site rather than having the replication complex move along the DNA.
Although the replication bubbles, with two daughter duplexes being made at each replication fork, are commonly used in replicating cellular DNA, other types of replicative structure have been found. For example, a type of replicative structure used by some bacteriophage to quickly generate many copies of the viral DNA is the rolling circle (Figure 5.5). A rolling circle is a replicative structure in which one strand of a circular duplex is used as a template for multiple rounds of replication, generating many copies of that template. When replication proceeds by a rolling circle, replication of one strand of the duplex begins at a nick at the origin. The newly synthesized strand displaces the original nicked strand, which does not serve as a template for new synthesis. Thus the rolling circle mechanism copies only one strand of the DNA. Elongation proceeds by the replication machinery going around the template multiple times, in a pattern resembling a rolling circle. The large number of copies of a single strand of a phage genome made by the rolling circle are concatenated, or connected end-to-end. The single‑stranded DNA can be cleaved and ligated to generate unit length genomes, which are packaged into phage particles. This occurs in replication of single‑stranded DNA phages such as fX174 or M13. The DNA in the bacteriophage particle is single stranded, and this strand is called the viral or plus strand. After infection of a bacterial cell the viral DNA is converted to a duplex replicative form, which is the double-stranded form of viral DNA used in replication. The new strand of DNA made during the conversion of the infecting single-stranded DNA to the replicative form is, of course, complementary to the viral strand, and it serves as the template during replication by the rolling circle mechanism. Thus the many copies of DNA produced are the viral strand, and these are packaged into viral particles. The rolling circle mechanism is not restricted to single-stranded bacteriophage. In some bacteriophage, the displaced single strand is subsequently copied into a daughter DNA duplex. The concatenated, multiple copies of genome‑length duplex DNA produced in this way are then cleaved into genome‑sized molecules and packaged into viruses. Thus the rolling circle mechanism followed by copying of the displaced strand can also be used to replicate some double-stranded phage. This occurs in the second phase of replication of bacteriophage l.
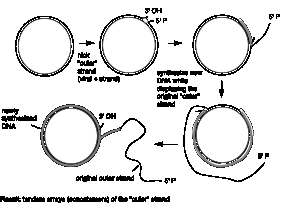

Figure 5.5. Rolling circles are structures formed as replication intermediates for some bacteriophage.
To further illustrate the range of replicative structures, consider a third structure, which is observed during replication of mitochondrial DNA (Fig. 5.6). Mitochondrial DNA synthesis starts at a specific, unidirectional origin on one strand. Initially only one of the parental strands is used as template for synthesis of a new strand. This single new strand displaces the non-template parental strand, forming a displacement loop, or D loop. After replication of the first strand has proceeded about half way round the mitochondrial genome, synthesis of the other strand begins at a second origin and proceeds around the genome.
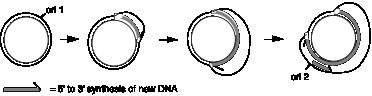
Figure 5.6. Displacement of D-loops in replication of mitochondrial DNA. Synthesis of one stand begins at an origin (ori1) and proceeds around the circular genome. The displaced strand is not immediately copied, and thus it forms a displacement loop, or D-loop. After synthesis from ori1 has proceeded about halfway around the genome, synthesis of the other strand begins at a second origin (ori2). The thick gray arrow is oriented with the tip of the arrowhead at the 3’ end of the newly synthesized DNA.
Question 5.2. How does D loop synthesis differ from a replication eye?
Although this is not an exhaustive list, these are three examples illustrate a range of replicative structures. Now we turn to a more detailed examination of events that occur at the replication fork of a molecule in which replication is proceeding via replication bubbles.
DNA synthesis at a replication fork of a replication bubble is semidiscontinuous.
In the common “eye-form” replication structure, or replication bubble, both daughter DNA molecules are synthesized at a replication fork. Because the two strands of DNA are antiparallel, one new strand must be synthesized in a 5’ to 3’ direction in the same direction as the fork moves, whereas the other strand must be synthesized in an overall 3’ to 5’ direction relative to fork movement (Fig. 5.7). One could imagine that this would occur by having two types of enzymes at the replication fork, one to catalyze synthesis in a 5’ to 3’ direction and another to catalyze synthesis in a 3’ to 5’ direction. Enzymes that catalyze the addition of deoxyribonucleotides to a growing chain of DNA are called DNA polymerases. Many of these have been isolated and studied, and all the DNA polymerases add nucleotides to a growing DNA chain exclusively in a 5' to 3' direction. Indeed, as will be discussed later, this polarity of synthesis is inherent in the chemical mechanism of DNA polymerization. So if no 3’ to 5’ synthesizing activity can be found, how is the new strand oriented 3’ to 5’ in the direction of the replication fork synthesized? The solution to this problem is discontinuous synthesis of that strand, whereas the other strand is synthesized continuously.

Figure 5.7. Semidiscontinuous DNA synthesis at the replication fork. The thick gray lines are newly synthesized DNA, and the arrows point toward the 3’ end.
As shown in Figure 5.7, one of the template DNA strands is oriented 3' to 5' at the replication fork, and hence it can be copied continuously by a DNA polymerase extending the new DNA chain in a 5’ to 3’ direction. This new DNA chain is called the leading strand; its orientation is 5' to 3' in the same direction as the fork movement. It is extending from the replication origin.
The other template strand is oriented 5’ to 3’ at the replication fork, and hence copying it will result in synthesis in a 3’ to 5’ direction relative to the direction of fork movement. This new DNA chain, called the lagging strand, is synthesized discontinuously, as a series of short DNA fragments. Each of these short DNA chains is synthesized in a 5’ to 3’ direction (right to left in Fig. 5.7, i.e. opposite to the direction of the replication fork). These short DNA fragments are subsequently joined together by DNA ligase to generate an uninterrupted strand of DNA. Because the leading strand is synthesized continuously and the lagging strand is synthesized discontinuously, the overall process is described as semidiscontinuous.
Okazaki and colleagues obtained evidence for discontinuous DNA synthesis during replication in 1968. Molecules in the process of being synthesized can be labeled by introducing radioactively labeled precursor molecules for a brief period of time; this procedure is called a pulse. For example, one can obtain the nucleoside thymidine labeled with tritium (3H). When this radiolabeled compound is added to the medium in which bacteria are growing, enzymes in the bacteria convert it to the nucleotide thymidine triphosphate, which is a precursor to DNA. The pulse period begins when the 3H thymidine is added to the medium, and it can be terminated by stopping cellular metabolism (for instance, by adding cyanide) in a process called quenching. Alternatively, the incorporation of labeled nucleotides can be ended by adding a large excess of unlabeled thymidine. When this is done, synthesis of the DNA continues during the remainder of the experiment, which is called a chase. The appearance of labeled nucleotides (incorporated during the pulse) in other parts of the product DNA can be monitored during the chase period. This latter design is particularly good for demonstrating that a given chemical intermediate is a precursor to a product.
Okazaki and colleagues used pulse labeling for increasing periods of time to examine DNA synthesis in bacteria. They labeled replicating DNA in E. coli with a brief pulse of [3H] thymidine ranging from 5 seconds to 5 minutes. They isolated the DNA and denatured it to separate the new (labeled) and old strands. The size of the newly replicated DNA was measured by sedimentation on denaturing sucrose gradients. At very short labeling times of 5 and 10 sec, most of the pulse-labeled DNA was small and sedimented slowly, with a sedimentation constant of about 10S corresponding to a size of about 1000 to 2000 nucleotides (Fig. 5.8). When the DNA was labeled for longer times (30 sec or greater), the amount of label in the short DNA segments reached a maximum whereas more and more label accumulated in larger, faster sedimenting DNA. The discrete population of short, newly synthesized DNA is evidence of discontinuous synthesis; a pulse-label of continuously synthesized DNA would have labeled large DNA molecules up to the size of the bacterial chromosome. (Even with the unavoidable shear of chromosomal DNA, the fragments would be much larger than the size of the small labeled DNAs.) The fact that larger DNA molecules are labeled at longer periods is indicates that the short fragments synthesized initially are subsequently joined together. We now understand that these small DNA segments are intermediates in discontinuous synthesis of the lagging strand, and they are called Okazaki fragments.
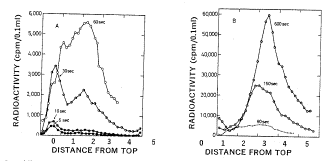
Figure 5.8. Incorporation of nucleotides into small DNA molecules during short labeling times demonstrates discontinuous synthesis of one DNA chain. Replicating DNA in E. coli was labeled with [3H] thymidine for the times indicated in the graphs, DNA was isolated from the cells, denatured and centrifuged on alkaline sucrose gradients to measure the size of the denatured DNA chains. Fast sedimenting, larger DNA is in the peaks toward the right. DNA labeled at short times sediments slowly (peak to the left) showing it is in the discrete small fragments, now called Okazaki fragments, that are intermediates in discontinuous synthesis of the lagging strand. This is Fig. 2 from R. Okazaki, T. Okazaki, K. Sakabe, K. Sugimoto, and A. Sugino (1968) Proceedings of the National Academy of Sciences, USA 59: 598-605.
Question 5.3. What would you expect to see if the replicating molecules were sedimented on a neutral (nonalkaline) sucrose gradient?
Enzymes and other components involved in replication can be identified biochemically and genetically.
The summary of activities shown in Fig. 5.7 suggests several types of enzymes that might be expected at the replication fork. Obviously, at least one DNA polymerase should be present, but we would also expect to find enzymes that unwind DNA, initiate the assembly of nucleotides, and join Okazaki fragments. Experiments that identified the components needed at the replication fork proceeded along two avenues – biochemical fractionation and genetic analysis. The detailed understanding of the mechanism of synthesis at the replication fork has come from both approaches, as well as the powerful combination of them in a technique called in vitro complementation.
Biochemical methods
Biochemical purification requires an assay for the activity under investigation, which is usually a measurement of the product of the reaction being catalyzed by the enzyme. Because the reaction catalyzed by DNA polymerases is the fundamental step in DNA replication, we will examine it in a little detail. A fairly simple way to observe the activity of a DNA polymerase is to measure the incorporation of a radioactively labeled deoxyribonucleoside or deoxyribonucleotide into the high molecular weight polymer DNA. The latter can be precipitated with a strong acid, such as trichloroacetic acid, whereas unincorporated nucleotides or nucleosides do not precipitate. For instance, a crude cell extract can be incubated with dTTP labeled with a 32P atom in the a-phosphate (abbreviated [a32P] dTTP) plus other unlabeled nucleoside triphosphates, appropriate buffers and cofactors. The DNA synthesized in this reaction can be measured as the amount of 32P precipitated by acid (Figure 5.9A).
The DNA polymerases can be separated from other macromolecules in the crude cell extract by a series of steps. Most (but not all) enzymes are proteins, and the procedures used in enzyme purification are primarily methods for separating proteins. For example, a researcher may separate a mixture of proteins on a series of chromatographic columns to separate proteins by charge, then by size, and then by hydrophobicity. Each fraction from a chromatography column is assayed for the DNA polymerase; the aim is to separate the proteins with the desired activity from as many other proteins as possible with each column (Figure 5.9B). The fractionation procedures are continued until the enzyme is purified, which usually means that only one polypeptide (or set of polypeptides for a multi-subunit protein) is detected by gel electrophoresis. In principle, it should be possible to isolate enzymes that can carry out any process for which a reliable assay is available. However, several factors can make such purification difficult, such as very low abundance of the desired protein in the starting material, or the need for a multi-subunit complex to carry out a reaction, especially if such a complex is not very stable.
![]() A.
A.

B. 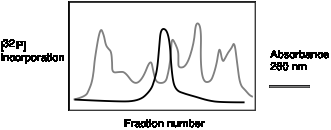
Figure 5.9. Biochemical assays for DNA polymerase. A. DNA synthesis can be assayed biochemically by the incorporation of a radioactive precursor, such as [a32P] dTTP, into acid-precipitable DNA as a function of time. B. Separation of a DNA polymerizing activity from other proteins by chromatography. Each fraction from a chromatographic column is assayed for total protein (e.g., the absorbance at 280 nm, gray line) and the ability to catalyze the incorporation of [a32P] dTTP into DNA (black line). In this hypothetical example, most of the proteins are in fractions other than the ones with the DNA polymerizing activity, and hence a substantial purification is achieved.
Many enzymes used in replication have been isolated by biochemical fractionation. These include not only the DNA polymerases, but also helicases, which unwind the parental DNA duplex to make two new templates, primase, which catalyzes the initial joining of nucleotides to start a DNA chain, DNA ligase, which joins fragments of DNA, and exonucleases, which can be used to remove incorrectly incorporated nucleotides. These and other enzymes have been used to reconstruct steps in DNA replication in the laboratory, and the activities described for these enzymes are used to build models for how replication can occur in living cells. Critical tests of such models can be made using genetic methods.
Genetic methods
Isolation and characterization of enzymes reveals proteins and RNAs that are capable of catalyzing reactions, and such activities can be used to postulate the events that occur in a biological pathway. Biochemical fractionation and analysis are rich sources of insight into the chemical reactions within cells and cellular physiology. However, such results do not necessarily tell us whether an enzyme purified using its ability to catalyze a reaction in the test tube is actually used to catalyze that reaction inside the cell. Such a conclusion is best made with genetic evidence.
A genetic analysis begins with a screen or a selection for mutants that are defective in the process under investigation. Of course, cells that are no longer able to synthesize DNA will not grow, so we must isolate conditional mutants. You should recall from Chapter 1 that the product of a conditional allele retains function under a permissive culture condition (e.g., low temperature, 33oC), but it loses activity at a restrictive culture condition (e.g., high temperature, 41oC). Other conditional mutants may be cold sensitive or salt sensitive. In the case of DNA synthesis, conditional mutants stop growing at the restrictive condition. Many temperature-sensitive mutants, i.e., those that do not grow at an elevated temperature such as 42 oC, were screened for the ability to synthesize DNA at the restrictive temperature. Such temperature-sensitive mutants in DNA synthesis were called dna mutants.
Once the conditional mutants have been isolated, they can be crossed to determine whether or not they complement at the restrictive temperature. Results of this analysis allows the mutants to be placed into complementation groups, where each complementation group represents a gene whose product is required for DNA synthesis in growing cells. The genes represented by these complementation groups are called dna genes, with each different gene given a different letter: dnaA, dnaB, etc. The aim of this genetic approach is to isolate a sufficiently large number of mutants such that at least one mutant is obtained in every gene needed for the process of interest, in this case DNA synthesis. If the genome were actually saturated with mutants, the number of complementation groups would be close to the number of genes encoding polypeptides carrying out the process under study. Studies in E. coli have revealed of the order of twenty dna genes. In order to find out what proteins and enzymatic activities are encoded by each of these genes, a method had to be developed to connect the genetically defined genes with a particular biochemical activity. This is the subject of the next section.
Combining genetic and biochemical methods
The method of isolating dna genes insures that their products are required for DNA synthesis. We would like to know exactly what enzymatic activity each gene encodes. For those activities for which a convenient in vitro assay is available, it is reasonably straightforward to find which mutants are defective in those activities at the restrictive condition. However, some dna genes may encode a protein with an activity that is not expected or readily assayed. The proteins can still be isolated using the powerful approach of in vitro complementation. This technique allows the isolation of an enzyme simply from the knowledge that a gene needed for replication encodes it. Rather than assaying for a particular enzymatic activity, one assays for the ability of an extract or chromatographic fraction to restore DNA synthesis in an extract of a temperature-sensitive dna mutant at the restrictive temperature.
As illustrated in Figure 5.10 A, cell extracts of dna mutants will not synthesize DNA at 41o (the restrictive temperature), but addition of an extract of wild-type cells will restore DNA synthesis in vitro. This in vitro complementation assay can be used to purify the protein from wild type extracts, assaying fractions from chromatographic columns for the ability to complement extracts from the temperature sensitive (abbreviated ts) cells (Fig. 5.10 B). Many of the products of the dna genes were isolated using this technique.
With this knowledge of the basic methods for identifying the enzymes needed for replication, we will proceed to a discussion of each of the major ones. We will cover DNA polymerases in considerable detail, whereas the other enzymes will be discussed less thoroughly.
A. B.

Figure 5.10. In vitro complementation to isolate enzymes needed for DNA replication. A. An extract of a temperature sensitive (ts) mutant defective in DNA synthesis cannot carry out this process at the restrictive temperature (dotted line). However, when a wild-type (wt) extract is added, synthesis is observed. This shows that the ts extract does not contain an inhibitor of synthesis, and thus a wild-type protein in vitro can complement it. B. The wild-type extract is fractionated by chromatographic methods, with each fraction assayed for the ability to restore in vitro DNA synthesis in the ts extract at the restrictive temperature. In this hypothetical illustration, the complementing activity (solid line) separates from many of the protein peaks (dotted line), showing a good purification at this step.
Studies of DNA polymerase I reveal essential information about the mechanism of polymerization.
Of all the enzymatic functions needed for replication of DNA, the ability to catalyze the incorporation of deoxynucleotides into DNA is most central. Enzymes that catalyze this reaction, DNA polymerases, have been isolated from many species, and many species have multiple DNA polymerases. Our earliest and most complete understanding of the mechanism of these enzymes comes from studies of the first DNA polymerase isolated, called DNA polymerase I.
Mechanism of nucleotide addition by DNA polymerases
In 1956 Arthur Kornberg and his co-workers isolated a protein from E. coli that has many of the properties expected for a DNA polymerase used in replication. In particular, it catalyzes synthesis of DNA from deoxynucleotides, it requires a template and it synthesizes the complement of the template. It is a single polypeptide chain of 928 amino acids, and it is the product of the polA gene. We now understand that this an abundant polymerase, but rather than synthesizing new DNA at the replication fork, it is used during the process of joining Okazaki fragments after synthesis and in DNA repair. Detailed studies of DNA polymerase I have been invaluable to our understanding of the mechanisms of polymerization. Although DNA polymerase I is not the replicative polymerase in E. coli, homologous enzymes are used in replication in other species. Also, the story of how the replicative DNA polymerases were detected in E. coli is a classic illustration of the power of combining biochemistry and genetics to achieve a more complete understanding of an important cellular process.
DNA polymerase I catalyzes the polymerization of dNTPs into DNA. This occurs by the addition of a dNTP (as dNMP) to the 3' end of a DNA chain, hence chain growth occurs in a 5' to 3' direction (Figure 5.11). In this reaction, the 3' hydroxyl at the end of the growing chain is a nucleophile, attacking the phosphorus atom in the a-phosphate of the incoming dNTP. The reaction proceeds by forming a phosphoester between the 3' end of the growing chain and the 5' phosphate of the incoming nucleotide, forming a phosphodiester linkage with the new nucleotide and liberating pyrophosphate (abbreviated PPi). Thus in this reaction, a phosphoanhydride bond in the dNTP is broken, and a phosphodiester is formed. The free energy change for breaking and forming these covalent bonds is slightly unfavorable for the reaction as shown. However, additional noncovalent interactions, such as hydrogen bonding of the new nucleotide to its complementary nucleotide and base-stacking interactions with neighboring nucleotides, contribute to make a total free energy change that is favorable to the reaction in the synthetic direction. Nevertheless, at high concentrations of pyrophosphate, the reaction can be reversed. In the reaction in the reverse direction, nucleotides are progressively removed and released as dNTP in a pyrophosphorolysis reaction. This is unlikely to be of large physiological significance, because a ubiquitous pyrophosphatase catalyzes the hydrolysis of the pyrophosphate to molecules of phosphate. This latter reaction is strongly favored thermodynamically in the direction of hydrolysis. Thus the combined reactions of adding a new nucleotide to a growing DNA chain and pyrophosphate hydrolysis insure that the overall reactions favors DNA synthesis. The basic chemistry of addition of nucleotides to a growing polynucleotide chain outlined in Fig. 5.11 is common to virtually all DNA and RNA polymerases.

Figure 5.11. Reaction catalyzed by DNA polymerases.
The DNA synthesis reaction catalyzed by DNA polymerase I requires Mg2+, which is a cofactor for catalysis, and the four deoxynucleoside triphosphates (dNTPs), which are the monomeric building blocks for the growing polymer. The reaction also requires a template strand of DNA to direct synthesis of the new strand, as predicted by the double helical model for DNA and confirmed by the Meselson and Stahl experiment.
This reaction also requires a primer, which is a molecule (usually a chain of DNA or RNA) that provides the 3’ hydroxyl to which the incoming nucleotide is added. DNA polymerases cannot start synthesis on a template by simply joining two nucleotides. Instead, they catalyze the addition of a dNTP to a pre-existing chain of nucleotides; this previously synthesized chain is the primer. The primer is complementary to the template, and the 3’ end of the primer binds to the enzyme at the active site for polymerization (Fig. 5.12). When a new DNA chain is being made, once a new nucleotide has been added to the growing chain, its 3' hydroxyl is now the end of the primer. The polymerase moves forward one nucleotide so that this new primer end is at the active site for polymerization. The alternative view, that the DNA primer-template moves while the DNA polymerase remains fixed, is also possible. In both cases the last nucleotide added is now the 3' end of the primer, and the next nucleotide on the template is ready to direct binding of another nucleoside triphosphate.
For the initial synthesis of the beginning of a new DNA chain, a primer has to be generated by a different enzyme; this will be discussed in more detail later in the chapter. For example, short oligoribonucleotides are the primers for the Okazaki fragments; these are found at the 5' ends of the Okazaki fragments and are made by an enzyme called primase. The RNA primers are removed and replaced with DNA (by DNA polymerase I) before ligation.
These requirements for Mg2+, deoxynucleotides and two types of DNA strands (template and primer) were discovered in studies of DNA polymerase I. We now realize that they are also required by all DNA polymerases.
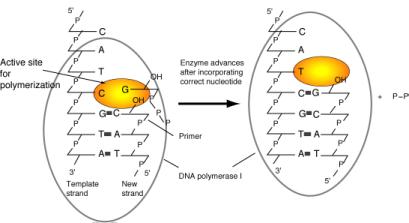
Figure 5.12. Chain elongation by DNA polymerase I. Binding of the correct incoming deoxynucleoside triphosphate to the active site for polymerization is directed by the deoxynucleotide on the template strand. The polymerase catalyzes formation of a phosphodiester bond with the new deoxynucleotide, and then it effectively moves forward so that the next deoxynucleotide on the template can direct binding of the next deoxynucleoside triphosphate to the active site. During elongation, the new DNA strand is also the primer.
The polymerization active site for DNA polymerase I has a specific dNTP-binding site (Fig. 5.12), and the active site adjusts to the deoxynucleotide on the template strand to favor binding of the complementary deoxynucleotide at the active site. Thus the polymerase catalyzes addition to the growing chain of the deoxynucleotide complementary to the deoxynucleotide in the template strand.
In the reaction catalyzed by DNA polymerase I, and all other DNA polymerases studied, the incoming deoxynucleotide is activated. The phosphoanhydride bonds in the triphosphate form of the deoxynucleotide are high-energy bonds (i.e., they have a negative, or favored, free energy of hydrolysis), and the b- and g- phosphates make a good leaving group (as pyrophosphate) after the nucleophilic attack. In contrast, the end of the growing DNA chain is not activated; it is a simple 3'-hydroxyl on the last deoxynucleotide added. This addition of an activated monomer to an unactivated growing polymer is called a tail-growth mechanism. DNA polymerases using this mechanism can only synthesize in a 5' to 3' direction, and all known DNA and RNA polymerases do this. Some other macromolecules, such as proteins, are made by a head-growth mechanism. In this case, the nonactivated end of a monomer attacks the activated end of the polymer. The lengthened chain again contains an activated head (from the last monomer added).
Question 5.4. Describe a hypothetical head-growth mechanism for DNA synthesis. In which direction does chain synthesis occur in this mechanism?
Proofreading the newly synthesized DNA
by a 3’ to 5’ exonuclease that is part of the DNA polymerase
The protein DNA polymerase I has additional enzymatic activities related to DNA synthesis. One, a 3’ to 5’ exonuclease, is intimately involved in the accuracy of replication. Nucleases are enzymes that catalyze the breakdown of DNA or RNA into smaller fragments and/or nucleotides. An exonuclease catalyzes cleavage of nucleotides from the end of a DNA or RNA polymer. An endonuclease catalyzes cutting within a DNA or RNA polymer. These two activities can be distinguished by the ability of an endonuclease, but not an exonuclease, to cut a circular substrate. A 3’ to 5’ exonuclease removes nucleotides from the 3’ end of a DNA or RNA molecule.
DNA synthesis must be highly accurate to insure that the genetic information is passed on to progeny largely unaltered. Bacteria such as E. coli can have a mutation rate, as low as one nucleotide substitution in about 109 to 1010 nucleotides. This low error frequency is accomplished by a strong preference of the polymerase for the nucleotide complementary to the template, which allows about one substitution every 104 to 105 nucleotides. The accuracy of DNA synthesis is enhanced by a proofreading function in the polymerase that removes incorrectly incorporated nucleotides at the end of the growing chain. With proofreading, the accuracy of DNA synthesis is improved by a factor of 102 to 103, so the combined effects of nucleotide discrimination at the polymerization active site plus proofreading allows only about one substitution in 106 to 108 nucleotides. Further reduction in the error rate is achieved by mismatch repair (Chapter 7).
The proofreading function of DNA polymerase I is carried out by a 3' to 5' exonuclease (Figure 5.13). It is located in a different region of the enzyme from the active site for polymerization. When an incorrect nucleotide is added to the 3' end of a growing chain, the rate of polymerization decreases greatly. The primer-template moves to a different active site on the enzyme, the one with the 3’ to 5’ exonucleolytic activity. The incorrect nucleotide is cleaved, and the primer-template moves back to the polymerization active site to resume synthesis. The enzyme distinguishes between correct and incorrect nucleotides at the 3’ end of the primer, such that the 3' to 5' exonuclease much more active when the terminus of the growing chain is not base paired correctly, but the polymerase activity exceeds that of the 3' to 5' exonuclease activity when the correct nucleotide is added.
The polymerizing activity and the proofreading 3' to 5' exonuclease found in DNA polymerase I are also found in most other DNA polymerases. These are central activities to DNA replication.
Tail growth mechanisms allow proofreading and subsequent
elongation. If the end of the
growing chain were activated (as in head growth), then proofreading would
eliminate the activated end and elongation could not continue.
D. A. C. B.
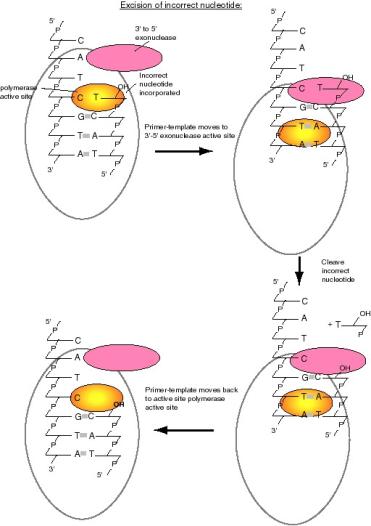
Figure 5.13. Excision of an incorrect nucleotide by DNA polymerase I. Incorporation an incorrect nucleotide (e.g., a T opposite a C, lower panel) (A) causes the primer-template to shift to the 3'-5' exonuclease active site (B) where the incorrect nucleotide is excised (C). The primer-template then can move back to the polymerase active site to resume synthesis (D).
Question 5.4 Removal of a nucleotide from the 3’ end of the growing chain by a 3’ to 5’ exonuclease is not the reverse of the polymerase reaction. Can you state what the difference is?
Removal
of nucleotides by a 5' to 3' exonuclease that is part of DNA polymerase I
In addition to the polymerase and 3’ to 5’ exonuclease common to most DNA polymerases, DNA polymerase I has an unusual 5' to 3' exonucleolytic activity. This enzyme catalyzes the removal of nucleotides in base-paired regions and can excise either DNA or RNA. It is used by the cell to remove RNA primers from Okazaki fragments and in repair of damaged DNA.
This 5' to 3' exonuclease, in combination with the polymerase, has useful applications in the laboratory. One common use is to label DNA in vitro by nick translation (Figure 5.14). In this process, DNA polymerase I will remove the DNA from a nicked strand by the 5' to 3' exonuclease, and then use the exposed 3' hydroxyl at the nick as a primer for new DNA synthesis by the 5' to 3' polymerase, thereby replacing the old DNA. The result is also a movement, or translation, of the nick from one point on the DNA to another, hence the process is called nick translation. If the reaction is carried out in the presence of one or more radiolabeled deoxynucleoside triphosphates (e.g., [a32P] dNTPs), then the new DNA will be radioactively labeled.
A similar process can be used to repair DNA in a cell. As will be discussed in Chapter 7, specific enzymes recognize a damaged nucleotide and cleave upstream of the damage. One way to remove the damaged DNA and replace it with the correct sequence is with the 5' to 3' exonuclease of DNA polymerase I and accompanying DNA synthesis.
![]()

[a32P]
Figure 5.14. The 5' to 3' exonuclease of DNA polymerase I can be used in nick translation to label DNA in vitro.
Structural
domains of DNA polymerase I
Further understanding of the mechanism of the three enzymatic functions of DNA polymerase can be obtained from a study of the three-dimensional (3-D) structure of the protein. Much of our knowledge of the structure of DNA polymerase I has come from biochemical characterization and more recently by determination of the 3-D structure using X-ray crystallography. These studies have shown that distinct structural domains of DNA polymerase I contain the different catalytic activities. Also, the 3-D structure provided the first look at what is now recognized as a common structure for many polymerases.
Mild treatment with the protease subtilisin cleaves DNA polymerase I into two fragments. The small fragment contains the 5' to 3' exonuclease, and the larger, or "Klenow," fragment (named for the biochemist who did the cleavage analysis) contains the polymerase and the proofreading 3' to 5' exonuclease (Figure 5.15). Thus the two activities common to most polymerases are together in the Klenow fragment, whereas the distinctive 5' to 3' exonuclease is in a separable domain. The fact that a mild treatment with a protease without a precise sequence specificity indicates that an exposed, readily cleaved domain connects the large and small fragments. Both these observations suggest that the 5' to 3' exonuclease was an active domain added to a polymerase plus proofreading domain during the evolution of E. coli. The Klenow polymerase is used in several applications in the laboratory, e.g., labeling the ends of restriction fragments by filling in the overhangs and sequencing by the dideoxynucleotide chain termination method.
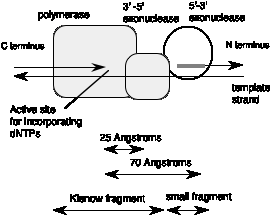
Figure 5.15. DNA polymerase I from E. coli has three active sites in three structural domains in one polypeptide.
The 3-D structure of the large fragment of DNA polymerase I, determined by crystallography, provides additional insight into the enzymatic functions of key structural components. The large fragment has a deep cleft, about 30 Ā deep, into which the template strand and primer bind. This cleft resembles a "cupped right hand" as illustrated in Fig. 5.16. The "palm" is formed by a series of b-sheets and the thumb and fingers are made by a-helices. The polymerase active site has been mapped within the deep cleft, with contributions from the b-sheets that form the palm and the a-helices forming the fingers. You can see more detailed views of the structure of the Klenow fragment at the Course/Book web site (currently http://www.bmb.psu.edu/courses/bmb400/default.htm. Click on the link to kinetic images, download the MAGE program and the kinemage file for DNA polymerase I, and view them on your own computer.)
The 3' to 5' proofreading exonuclease is located in another part of the structure of the Klenow fragment, about 25 Ā from the polymerase active site. Thus the primer terminus has to move this distance in order for the enzyme to remove misincorporated nucleotides.
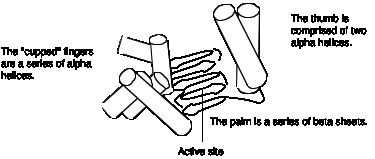
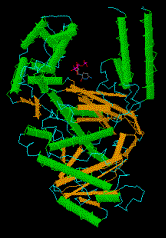
Figure 5.16. The portion of DNA polymerase I used for DNA synthesis resembles a "cupped right hand." A view of the structure with dCTP bound to the active site, rendered by CN3D from the NCBI web site (MMDB Id: 1395 PDB Id: 1KFD), is shown below the drawing. We may want to add a copy of panel A of Fig. 2 of Kim et al. (1995) Crystal structure of Thermus aquaticus DNA polymerase, Nature 376:612-616, which shows all 3 domains.
The large Klenow fragment of the E. coli DNA polymerase I lacks the 5’ to 3’ exonuclease, so the 3-D structure of the Klenow fragment gives no information about that exonuclease. However, the 5’ to 3’ exonuclease domain can be seen in the structure of DNA polymerase from the thermophilic bacterium Thermus aquaticus. This protein structure is very similar to that of DNA polymerase I of E. coli in the polymerase and 3’ to 5’ exonuclease domains, and it has an additional 5' to 3' exonuclease domain located about 70 Ā from the polymerase active site. This is a large distance, but remember that this exonuclease is working on a different region of the DNA molecule than the polymerase. The 5’ to 3’ exonuclease uses one part of the DNA molecule as a substrate for excising primers or removing damaged DNA, whereas the polymerase uses a different part of the DNA molecule as a template to direct synthesis of a new strand.
Curiously, a region homologous to the proofreading 3' to 5' exonuclease domain of DNA polymerase I is present in the Thermus aquaticus polymerase structure, but it is no longer functional. The absence of proofreading accounts for the elevated error rate in this polymerase used very commonly for amplification of DNA by PCR. Of course, this polymerase is used in PCR because it is stable at the high temperatures encountered during the cycles of PCR. Some other thermostable polymerases with a lower error rate have become available more recently for use in PCR.
Similar "cupped right hand" structures occur in the tertiary structure of T7 RNA polymerase and the HIV reverse transcriptase. Thus DNA polymerase I was the first member described in what we now realize is a large class of nucleic acid polymerases. This family includes single unit polymerases for both RNA and DNA synthesis. You can access a tutorial on the T7 DNA polymerase at http://www.clunet.edu/BioDev/omm/exhibits.htm#displays. This structure has some similarities to that of DNA polymerase I.
Physiological role of DNA polymerase I
Although studies of DNA polymerase I have provided much information about the mechanism of DNA synthesis, genetic analysis has shown that the polymerase function of this enzyme is not required for DNA replication. DNA polymerase I is encoded by the polA gene in E. coli. However, no mutant allele of polA was isolated in screens for conditional mutants defective in DNA replication. The most compelling argument that this polymerase is not required for replication came from an examination of thousands of E. coli mutants, assaying them for DNA polymerase I activity. A mutant polA strain was isolated (Fig. 5.16). This mutant allele, called polA1, contained a nonsense codon, leading to premature termination of synthesis of the product polypeptide and hence a loss of polymerase function. However, the mutant strain grew at a normal rate, which shows that DNA polymerase I is not required for DNA synthesis. The most striking phenotype of the polA1 mutant was its strongly reduced ability to repair DNA damage. Further investigation led to the isolation of conditional lethal alleles of the polA gene. The mutant DNA polymerase I proteins encoded by these conditional lethal alleles are defective in the 5' to 3' exonuclease activity, demonstrating that this activity is required for cell viability. The 5' to 3' exonuclease activity removes RNA primers during synthesis of the lagging strand at the replication fork, and it is used in DNA repair.
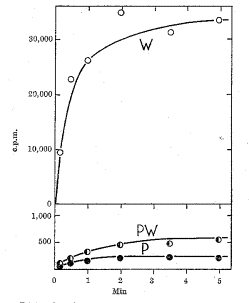
Figure 5.16. Extracts from a polA1 mutant strain are defective in DNA polymerase activity. DNA polymerization in extracts of E. coli cells was measured by the incorporation of radiolabeled dTTP into DNA. The wild type strain (line labeled W) showed high activity. Mutants of this strain were systematically screened for the loss of this DNA polymerizing activity, and one was found (line labeled P; note the change in scale from the upper panel). This mutant strain has less than 1% of the wild type activity, as shown by the mixing 1 part of wild-type extract with 99 parts of the mutant extract (a 100-fold dilution; results are shown as line PW). The mutation was mapped to polA, which encodes DNA polymerase I. The mutant strain grows as well as the wild type, showing that DNA polymerase I is not required for DNA replication. This figure is from De Lucia and Cairns (1969) Nature 224:1164-1166.
DNA polymerase III is a highly
processive, replicative polymerase.
The conclusion that DNA polymerase I is not the replicative polymerase for E. coli led to the obvious question of what enzyme is actually used during replication. Investigation of the genes isolated in screens for mutants that are conditionally deficient in replication led to the answer. The replicative polymerase in E. coli is DNA polymerase III.
DNA polymerase I is more abundant than other polymerases in E. coli and obscures their activity. Thus the depletion of DNA polymerase I activity in polA1 mutant cells (Fig. 5.17) provided the opportunity to observe the other DNA polymerases. DNA polymerases II and III were isolated from extracts of polA1 cells, named in the order of their discovery.
DNA polymerase II is a single polypeptide chain whose function is uncertain. Strains having a mutated gene for DNA polymerase II (polB1) show no defect in growth or replication. However, the activity of DNA polymerase II is increased during induced repair of DNA, and it may function to synthesize DNA opposite a deleted base on the template strand.
Genetic evidence clearly shows that DNA polymerase III is used to replicate the E. coli chromosome. This enzyme is composed of multiple polypeptide subunits. Several of the genes encoding these polypeptide subunits were identified in screens for conditional lethal mutants defective in DNA replication. Loss of function of these dna genes blocks replication, showing that their products are required for replication.
Low abundance and high processivity of
DNA polymerase III
DNA polymerase III has many of the properties expected for a replicative polymerase. One of the complications to studies of DNA polymerase III is that different forms were isolated by various procedures. We now realize that these forms differ in the number of subunits present in the isolated enzyme. For enzymes with multiple subunits, we refer to the complex with all the subunits needed for its major function as the holoenzyme or holocomplex. The DNA polymerase holoenzyme has ten subunits, which will be discussed in detail in the next section.
It is the DNA polymerase holoenzyme that has the properties expected for a replicative polymerase, whereas DNA polymerase I does not (see comparison in Table 5.1). It is less abundant than DNA polymerase I, but large number of replicative DNA polymerases are not needed in the cell. Only one or two polymerases can be used at each replication fork, so the 10 molecules of the DNA polymerase III holoenzyme will suffice. DNA polymerase III catalyzes DNA synthesis at a considerably higher rate than DNA polymerase I, by a factor of about 70. The elongation rate measured for the DNA polymerase III holoenzyme (42,000 nucleotides per min) is close to the rate of replication fork movement measured in vivo in E. coli (60,000 nucleotides per min).
A key property for a replicative DNA polymerase is high processivity, which is a striking characteristic of the DNA polymerase III holoenzyme. Processivity is the amount of polymerization catalyzed by an enzyme each time it binds to an appropriate template, or primer-template in the case of DNA polymerases. It is measured in nucleotides polymerized per binding event. In order to replicate the 4.5 megabase chromosome of E. coli in 30 to 40 min, DNA polymerase needs to synthesize DNA rapidly, and in a highly processive manner. DNA polymerase I synthesizes less than 200 nucleotides per binding event, but as the holoenzyme, DNA polymerase III is much more processive, exceeding the limits of the assay used to obtain the results summarized in Table 5.1. In contrast, the DNA polymerase III core, which has only three subunits (see next section), has very low processivity.
Table 5.1. Comparison of DNA polymerases I and III (Pol I and Pol III)
|
Property |
Pol I |
Pol III core |
Pol III holoenzyme |
|
molecules per cell |
400 |
40 |
10 |
|
nucleotides polymerized min-1 (molecule enzyme)-1 |
600 |
9000 |
42,000 |
|
processivity [nucleotides polymerized per initiation] |
3-188 |
10 |
>105 |
|
5' to 3' polymerase |
+ |
+ |
+ |
|
3' to 5' exonuclease, proofreading |
+ |
+ |
+ |
|
5' to 3' exonuclease |
+ |
- |
- |
Note: + and – refer to the presence or absence of the stated activity in the enzyme.
Question 5.6. If the
rate of replication fork movement measured in vivo in E. coli is 60,000 nucleotides per min, how many forks are
needed to replicate the chromosome in 40 min? Recall that the size of the E.
coli chromosome is 4.64 ę 106 bp.
Subunits and
mechanism of DNA polymerase III
The DNA polymerase III enzyme has four distinct functional components, and several of these contain multiple subunits, as listed in Table 5.2 and illustrated in Figure 5.18. The a and e subunits contain the major polymerizing and proofreading activities, respectively. They combine with the q subunit to form the catalytic core of the polymerase. This core can be dimerized by the t2 linker protein to form a subassembly called DNA polymerase III'. Addition of the third functional component, the g complex, generates another subassembly denoted DNA polymerase III*. All of these subassemblies have been isolated from E. coli and have been characterized extensively. The final component is the b2 dimer, which when combined with DNA polymerase III* forms the holoenzyme.
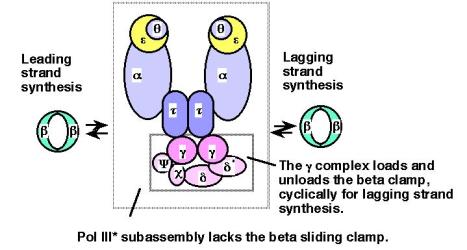
Figure 5.18. Subunits and subassemblies of DNA polymerase III. The a, e, and q subunits form the catalytic core, and two t subunits are thought to hold the two cores in a single large complex. One g complex is present in the DNA polymerase III* subassembly and in the holoenzyme, forming an asymmetric dimer for the overall structure. Addition of the ring-shaped b2 dimers greatly increases processivity. The arrows between the b2 dimers and the PolIII* subassembly denote the cyclical addition and removal of this subunit during synthesis, which is explored more in more detail in Fig. 5.19.
The various activities of DNA polymerase III can be assigned to individual subunits (Table 5.2). For instance, the major polymerase is in the a subunit, which is encoded by the dnaE gene (also known as polC). The 3' to 5' exonuclease is in the e subunit, which is encoded by the dnaQ gene (also known as the mutD gene). However, maximal activity is obtained with combinations of subunits. The DNA polymerase III core is a complex of the a, e and q subunits, and the activity of the core in both polymerase and 3' to 5' exonuclease assays is higher in than in the isolated subunits.
Table 5.2. Subassemblies of DNA polymerase III, major subunits, genes and functions
|
Functional component |
Subunit |
Mass (kDa) |
Gene |
Activity or function |
|
Core polymerase |
a |
129.9 |
polC=dnaE |
5' to 3' polymerase |
|
|
e |
27.5 |
dnaQ=mutD |
3'-5' exonuclease |
|
|
q |
8.6 |
|
Stimulates e exonuclease |
|
Linker protein |
t |
71.1 |
dnaX |
Dimerizes cores |
|
Clamp loader |
g |
47.5 |
dnaX |
Binds ATP |
|
(or g complex) |
d |
38.7 |
|
Binds to b |
|
(ATPase) |
d' |
36.9 |
|
Binds to g and b |
|
|
c |
16.6 |
|
Binds to SSB |
|
|
y |
15.2 |
|
Binds to c and g |
|
Sliding clamp |
b |
40.6 |
dnaN |
Processivity factor |
The activities of
the subunits can be measured in vitro by
appropriate biochemical assays. In addition, the phenotype of mutations in the
gene encoding a given subunit can show that subunit is required for a
particular process. Mutant a
subunits are the product of conditional lethal alleles discovered in screens
for dna genes, but they also were
discovered as the product of polymerase-defective alleles defining the polC gene. Thus the dnaE gene is the same as same as the polC gene, showing that this subunit with polymerase
activity is needed in replication. Similarly, the phenotype of mutations in the
gene encoding the e subunit
shows that it is needed for proofreading. Mutant alleles of the dnaQ gene were identified in a screen for mutator
genes, which generate a high frequency of mutants in bacteria when defective.
These alleles defined a gene mutD, which
was subsequently shown to be the same as dnaQ. The mutator phenotype of mutant dnaQ/mutD strains results from a lack of proofreading by the e subunit during replication, allowing more
frequent incorporation of incorrect nucleotides into DNA.
The b2 dimer is the key protein that confers high processivity on DNA polymerase III. Association of the b2 dimer with DNA polymerase III increases the processivity from about 10 nucleotides polymerized per binding event to over 100,000 nucleotides polymerized per binding event (Table 5.1). This dimeric protein forms a ring through which the duplex DNA can pass; the ring will slide easily along DNA unless impeded, as, for example, by proteins bound to the template DNA. Thus the b2 dimer acts as a sliding clamp, holding the polymerase onto the DNA being copied. Once DNA polymerase III is associated with the clamp on DNA, it will polymerize until it reaches the next primer for an Okazaki fragment during lagging strand synthesis. For leading strand synthesis, the DNA polymerase presumably remains associated with the DNA via the b2 clamp until the chromosomal DNA is completely replicated. The 3-D structure of the b2 dimer, determined by X-ray crystallography, shows a macromolecular ring. This structure can be viewed at the web site for the course and at the Online Museum of Macromolecules (http://www.clunet.edu/BioDev/omm/exhibits.htm#displays).
The g-complex contains several subunits: two molecules of g subunits and one molecule each of d, d', c, and y. It loads the b2 dimer clamp onto a primer-template, in a process that requires ATP hydrolysis (Figure 5.19). The catalytic core of DNA polymerase III will then link to the template-bound clamp and will initiate highly processive replication. The g-complex also serves to unload the clamp once an Okazaki fragment is completed during lagging strand synthesis; hence it is both a clamp loader and unloader, allowing the polymerase and the clamp to cycle repeatedly from one Okazaki fragment to another.
The g-complex carries out these opposite activities on different structures, loading on the clamp at a template-primer and unloading the clamp at the end of a completed Okazaki fragment. For instance, encountering the 5' end of the previously synthesized Okazaki fragment may be the distinctive structure that shifts the g-complex into its unloading mode. It does not unload the clamp while DNA polymerase III is catalyzing polymerization.
Figure 5.19
illustrates the proposed steps in this process. The g-complex in the ATP-bound form binds the b2 clamp, whereas the g-complex in the ADP-bound form releases the b2 clamp. Thus loading and
unloading depend on a round of ATP hydrolysis. When the g-complex in the ATP-bound form binds the b2 clamp, the DNA polymerase III
holoenzyme is in a conformation that allows it to find a primer-template. The
ring of the b2 clamp is held
open by the g-complex-ATP, allowing it
to bind around a primer-template. Hydrolysis of ATP by the g-complex leaves it in an ADP-bound form. In
this new, ADP-bound conformation of the g-complex,
it dissociates from the b2
clamp, thereby allowing the b2
clamp to bind to the catalytic core of the holoenzyme and also close around the
primer-template. The holoenzyme is now ready to catalyze processive DNA
synthesis. Elongation continues until the holoenzyme encounters a previously
synthesized Okazaki fragment. Now the g-complex
binds ATP (presumably by an ADP-ATP exchange reaction) and shifts into the
conformation for binding to the b2
clamp and taking it off the DNA template. This half of the holoenzyme is now
able to dissociate from the template and find the next primer-template junction
to begin synthesis of another Okazaki fragment.
The
clamp loading and unloading activities of the g-complex
are a cycle of changes in protein associations. These changes occur because of
the enzymatic activities of the complex, which in turn alter the conformations
of the proteins and their preferred interactions. As shown in Fig. 5.19, the g-complex is an ATPase, which is an enzyme that catalyzes the hydrolysis
of ATP to ADP and phosphate. It is also an ATP-ADP exchange factor.
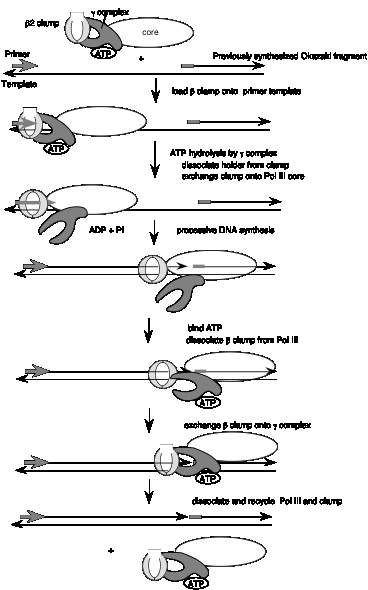
Figure 5.19. (Previous page) Loading and unloading of the b2 clamp by the g-complex in the DNA polymerase III holoenzyme. The b2 clamp is shown as a ring that can be bound and opened by the ATP-bound form of the g-complex (shown as a pincher shape, but its shape is not known). After initial binding of the b2 clamp, ATP-hydrolysis by the g-complex causes this complex to shift into an ADP-bound conformation that allows it to release the b2 clamp, so that the b2 clamp is loaded onto the primer template and also linked to the catalytic core of the polymerase. After completion of the Okazaki fragment, the g-complex exchanges the ADP for ATP, and is now is a conformation to bind the b2 clamp and unload it. Not all the steps in this model have been demonstrated, but it is useful to illustrate how cycles of ATP hydrolysis could be used in loading and unloading the b2 clamp. The figure shows only the half of the DNA polymerase III holoenzyme engaged in synthesis of the lagging strand; the other half is thought to be engaged in synthesis of the leading strand, but is not shown here to keep the diagram relatively simple.
Changes in conformation and activity of proteins depending on whether they are bound to a nucleoside triphosphate (ATP or GTP) or a nucleoside diphosphate (ADP or GDP) is a common theme in biochemistry. The GTP-bound forms of proteins, which can be turned off by GTP-hydrolysis and reactivated by GDP-GTP exchange proteins, mediate critical cell signaling events. As will be seen in Chapter 14, GTP- and GDP-bound forms of translation factors carry out opposite functions. Proteins assume different conformations depending on the cofactor bound (in this case a nucleotide), and each conformation has a distinct activity. The ability to change the conformation by a hydrolytic activity (converting ATP to ADP and phosphate) allows the protein to shift activities readily.
The two catalytic cores of DNA polymerase III are joined together by the t subunits to make an asymmetric dimer (see Figure 5.18). The half of the holoenzyme without the g complex is proposed to synthesize the leading strand of new DNA, and the core with the g complex is proposed to synthesize the lagging strand. Both of the cores in the asymmetric dimer are associated with a b2 clamp at the replication fork. In this model, synthesis of both the leading and lagging strands is catalyzed by the same DNA polymerase III complex, thereby coordinating synthesis of both new strands strand. Note that if the template for lagging strand synthesis is looped around the enzyme, then leading and lagging strand synthesis would be occurring in the same direction as replication fork movement (Figure 5.20), despite the opposite polarities of the two template strands. Thus the asymmetric dimer model suggests a means to couple both leading strand and lagging strand synthesis.
 Replication machinery
Replication machinery
C. B. A.
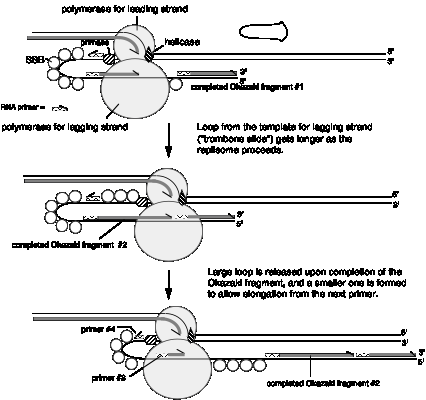
Figure 5.20. Simultaneous synthesis of both leading and lagging strands by an asymmetric dimer of DNA polymerase III. In this model, one of the catalytic cores synthesizes the leading strand and the other synthesizes the lagging strand. The template for the lagging strand may be looped around the polymerase, making this strand resemble the slide on a trombone (panel A); this model has been called the Trombone Slide model. When this is done, the synthesis of the Okazaki fragments is in the same direction as leading strand synthesis and fork movement (i.e. left to right in the diagram). Effectively, wrapping the template strand for lagging strand synthesis around the polymerase orients this strand at the active site in the same polarity as the template for leading strand synthesis. (A) The enzyme primase makes a primer in the enlarging single stranded loop, while the DNA polymerase III core catalyzes extension of the Okazaki fragment. (B) The Okazaki fragment is completed, and DNA polymerase III encounters the previously synthesized Okazaki fragment. (C) Now the loop with the completed Okazaki fragment is released and a new single stranded loop is formed. DNA polymerase III initiates replication at the primer in the new loop, and lagging strand replication resumes, again moving in the same direction as the fork (and the leading strand). SSB is single-strand binding protein and primase catalyzes the synthesis of primers (which are mainly RNA) for Okazaki fragments; these will be discussed in more detail later.
Proteins in addition to DNA polymerases are needed for replication.
The assembly of nucleotides into a polymer in a template-directed manner, catalyzed by DNA polymerases, is the core activity of replication. However, it is not sufficient. Several additional enzymes are needed (Fig. 5.21). In this section, we will discuss DNA helicases, topoisomerases, primases, and ligases. Some of these have been encountered in earlier chapters with regard to their use in recombinant DNA techniques and in investigations of DNA topology. The primase does not work as a single polypeptide, and its function within a complex called a primosome will also be covered.
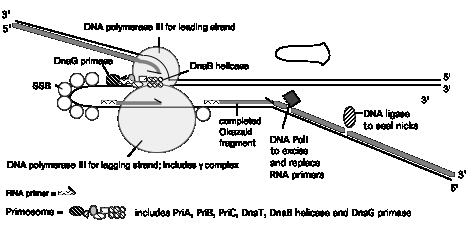
Figure 5.21. Summary of replication fork movement in E. coli. Helicases such as DnaB and PriA unwind the parental duplex, and SSB stabilizes the single strands. Topoisomerases provide a swivel to prevent an excessive accumulation of supercoils in the DNA. Continuous synthesis on the leading strand is catalyzed by one of the catalytic cores of the DNA polymerase III holoenzyme, held on to the template by the b2 sliding clamp for high processivity. The primer for synthesis of the leading strand is made during initiation (see Chapter 6). Discontinuous synthesis on lagging strand requires more proteins. The seven proteins PriA, PriB, PriC, DnaT, DnaC, DnaB, and DnaG (primase) are used in assembling the primosome, and DnaG makes primers at appropriate places, directed by the template for lagging strand synthesis. The other catalytic core of the DNA polymerase III holoenzyme makes a new Okazaki fragment, extending from the primer to the previously synthesized Okazaki fragment. The 5' to 3' exonucleolytic activity of DNA polymerase I removes the primers, and the polymerase activity of this same enzyme can fill in the resulting gap in the new DNA. DNA ligase then seals the nicks left between the Okazaki fragments, producing a continuous DNA strand from the Okazaki fragments.
Changes in DNA topology during
replication
The two strands of the parental DNA helix must be unwound in order for the polymerase to read each template and synthesize the new complementary strand. Enzymes that catalyze this separation are called DNA helicases. They catalyze the unwinding of the DNA duplex as the replication fork moves, usually using the energy of ATP hydrolysis to drive the process. Two molecules of ATP are hydrolyzed for each base pair that is unwound. Their activity can be measured biochemically by the conversion of duplex DNA to single-stranded DNA.
DNA helicases also have a second activity. They can move along single stranded DNA with a specific polarity. The polarity of movement can be measured by an in vitro assay using a substrate in which two distinctive short, labeled strands are in duplex with the two ends of a longer strand (Figure 5.21). A helicase will bind initially to the single-stranded portion of the substrate DNA and track along it until it meets a duplex region, at which point it will catalyze the unwinding of the duplex. Only one or the other of the short, labeled strands will be displaced by the helicase, depending on the direction of tracking along the single-stranded region. The displacement of molecule A in the Figure 5.22A shows that this helicase (DnaB) moved in a 5' to 3' direction along the single stranded DNA to reach the duplex portion including A. Fragment A was then displaced by the helicase activity.
A. B.
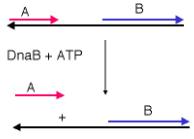
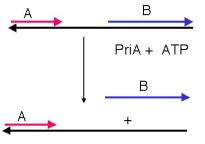
Figure 5.22. An assay for direction of helicase tracking along a single stranded region.
Question 5.7. Figure 5.22B shows the results of the tracking assay for a helicase called PriA. In what direction does it track along the single-stranded DNA?
Seven helicases have been isolated from E. coli. A distinctive function has not been defined for all of them, nor is it completely clear which act at the replication fork. The principal helicases for E. coli chromosomal replication appear to be PriA and DnaB, which are also used in the machinery for making primers. An additional helicase that will be considered in Chapter 7 is helicase II, or UvrD, used in repair of DNA.
Once the two strands of the parental DNA molecule have been separated, they must be prevented from reannealing. Coating the single-stranded DNA with single-stranded DNA binding protein (SSB) does this (Figure 5.21). SSB from E. coli is a homotetramer, which a complex of four identical monomeric subunits. The monomeric protein subunits are 74 kDa; they are encoded by the ssb gene. Mutants in ssb stop DNA synthesis immediately, and hence they are needed for elongation. Such loss-of-function mutants are also defective in repair and recombination of DNA. SSB binds cooperatively to single‑stranded DNA, and in this form the single-stranded DNA cannot anneal to its complementary strand. An analogous protein found in eukaryotes is Replication Factor A, or RFA. This is a heterotrimer, i.e., composed of three different subunits.
The unwinding of the DNA strands by helicases affects the overall topology of the DNA (Fig. 5.23). For instance, for every 10 base pairs that are unwound (DT=-1), there will be a compensatory increase in writhing (DW=+1) unless the linking number is changed. As discussed earlier in Chapter 2, enzymes that can change the linking number are topoisomerases. These enzymes act as swivels during replication to relieve the topological strain. In E. coli, this swivel is the enzyme gyrase (Table 5.3, Fig. 5.23). This topoisomerase II uses the energy of ATP to make a double-strand break in a DNA molecule, passes a different portion of the DNA through the break, and reseals the break. The direction of passage is such that a negative superhelical turn is introduced, thereby counteracting the positive change in W that occurs when DNA is unwound. Compounds that inhibit gyrase, such as naladixic acid or the coumarins, also block DNA synthesis, showing the critical role for gyrase in this process. In mammalian cells, either topoisomerase I or topoisomerase II can be used as the swivel. Topoisomerase I makes a single stranded nick in supercoiled DNA, thereby allowing the supercoils to relax.
Fig. 5.23. Changes
in topology of the DNA during replication. The ATP-dependent untwisting
(negative DT) catalyzed by DNA
helicases causes a compensatory change in writhing (positive DW), which is relieved by the action of a
topoisomerase II, such as DNA gyrase. Gyrase catalyzes an ATP-dependent, three-step
reaction, cleaving the two strands of DNA, passing another part of the DNA
duplex through the break and re-sealing the break. The action of gyrase
generates a negative DW to balance the
positive DW from the action of
helicase. X and Y mark two different regions of the DNA molecule. A gray arrow
indicates the direction of duplex movement through the break. Gyrase is a
tetramer, and is shown as four pink balls.
Making primers for DNA synthesis
The enzyme primase catalyzes the synthesis of the primers from which DNA polymerases can begin synthesis (Figure 5.21). Primers are short oligonucleotides, ranging from 6 to 60 nucleotides long. They can be made of ribonucleotides or a mixture of deoxyribonucleotides and ribonucleotides. The principal primase in E. coli is the 60 kDa protein called DnaG protein, the product of the dnaG gene. The major primase in eukaryotic cells is DNA polymerase a.
The primase of E. coli, DnaG protein, cannot synthesize primers by itself, but rather it is part of much larger complex called the primosome. The primosome acts repeatedly during lagging strand synthesis, finding a primer-binding site on the SSB-coated single-stranded template strand and synthesizing a primer. Identification of the components of the primosome was aided by the convenient model system of in vitro synthesis of fX174 DNA. fX174 is a single-stranded bacteriophage; the DNA found in the virus is termed the plus strand. After infection of E. coli, this plus strand is converted to a double-stranded replicative form (Figure 5.24 A). The conversion of single‑stranded phage DNA to duplex DNA occurs by the synthesis of several Okazaki fragments, and hence it is a good model for discontinuous synthesis on the lagging strand. This reaction can be carried out in vitro, which allowed the biochemical dissection of the various steps in primosome assembly and movement.
Table 5.3. Components of the primosome in E. coli
|
protein |
gene |
activities and functions |
|
PriA |
priA |
helicase, 3' to 5' movement, site recognition |
|
PriB |
priB |
|
|
PriC |
priC |
|
|
DnaT |
dnaT |
needed to add DnaB‑DnaC complex to preprimosome |
|
DnaC |
dnaC |
forms complex with DnaB |
|
DnaB |
dnaB |
helicase, 5' to 3' movement. DNA dependent ATPase. |
|
DnaG |
dnaG |
synthesize primer |
Five different proteins are found in a prepriming complex, PriA, PriB, PriC, DnaT, and DnaB (Table 5.4). A sixth protein, DnaC, is needed for the assembly of this complex. In the case of fX174 viral DNA template coated with SSB, PriA (Figure 5.24 B) recognizes a primer assembly site. The proteins PriB and PriC are then added to form a complex. The hexameric protein DnaB is in a complex with six molecules of DnaC when it is not on the DNA. In an ATP-dependent process, and with help from DnaT, DnaB is transferred to the template and DnaC is released.
The prepriming complex is now ready for the primase, DnaG, to bind and make the active primosome. Although the role of each of the proteins in the primosome is not yet clear, information is available on some of the steps in primosome action. The preferred binding site on the template for primase is CTG, with the T being used as the template for the first nucleotide of the primers. A high affinity complex between DnaB and ATP forms or stabilizes a secondary structure in the single-stranded template DNA that is used by primase; this is thought to be how DnaB "activates" the primase to begin synthesis. After ATP hydrolysis by DnaB, the low affinity ADP-DnaB complex dissociates from the template. The primosome can now move to the next site for primer synthesis.
A. B.
Figure 5.24. Assembly and migration of the primosome. After assembly of the prepriming complex, DnaG joins the complex to complete the primosome. After synthesis of the primer (dark black line), the primosome moves to the next site for synthesis. This tracking along the SSB-coated single stranded DNA requires ATP hydrolysis and causes dissociation of some of the SSB. In the diagram, the primosome is shown moving in a 5' to 3' direction along the template strand (clockwise), which is the opposite of the direction of primer synthesis. This would be the direction of movement catalyzed by DnaB. The primosome also contains PriA, which catalyzes movement along single-stranded DNA in the opposite direction. Once primers have been synthesized, DNA polymerase III can synthesize Okazaki fragments from them, the primers are excised and gaps repaired by DNA polymerase I, and then the fragments are ligated together.
The primosome contains two helicases than can move along single-stranded DNA with opposite polarity. PriA moves in a 3' to 5' direction, whereas DnaB moves in a 5' to 3' direction. When tested in vitro with a substrate similar to that shown in Figure 5.22, fragments from each end are displaced, indicating that the primosome moved in one direction on some molecules and in the other direction on others. Figure 5.24 B shows the migration as driven by the DnaB helicase, but movement can also occur in the other direction as well.
Removal of RNA primers and joining Okazaki fragments
Once a series of short Okazaki fragments has been made during discontinuous DNA synthesis of the lagging strand, the primers are excised by DNA polymerase I. This polymerase also copies the primer template into DNA to insure that only nicks are left between the Okazaki fragments. These nicks can then be sealed by DNA ligase, which catalyzes the covalent joining of the fragments into a long DNA strand. Catalysis by DNA ligase occurs in two stages (Figure 5.25). First, the enzyme is modified by adenylylation to make an active intermediate. The AMP can come from NAD (in the case of E. coli DNA ligase) or ATP (in the case of T4 DNA ligase). (The T4 DNA ligase is widely used to make recombinant DNA molecules in vitro.) This active enzyme intermediate then transfers the AMP to the 5' phosphate at the nick in the substrate DNA, thereby activating it. The 3' hydroxyl at the nick then attacks the adenylylated 5' end of the chain, forming the new phosphodiester bond to seal the nick and releasing AMP.

Figure 5.25. Mechanism
of action for DNA ligase. In the first of two steps, the enzyme is adenylylated
to form the active intermediate. In the second step, this enzyme intermediate
activates the 5' phosphate at the nick in the substrate DNA by transferring the
AMP to it. The 3' hydroxyl at the nick is a nucleophile that attacks the
adenylylated 5' end of the chain, forming a new phosphodiester bond and
releasing AMP.
Replisome
It is plausible that the multisubunit primosome is associated with the DNA polymerase III holocomplex in an even larger complex called a replisome at the replication fork. DnaB and PriA are the primary helicases in replication, and thus the proteins in the primosome may also be functioning to unwind and move DNA strands at the replication fork. DnaB-directed movement of the primosome in the 5' to 3' direction along the template for lagging strand synthesis corresponds to the direction of replication fork movement (Figure 5.26). In contrast, movement directed by PriA would be in the opposite direction. These opposite activities can be accommodated in a "sewing machine" model for the replication fork, in which the replisome is postulated to be stationary (the sewing machine) and the DNA being copied is moved through it, much like moving cloth through a sewing machine. If DnaB is at the front end of the replisome, it can unwind the parental duplex DNA and pull the template for lagging strand synthesis into a loop (Figure 5.26). The DNA tracking activity of PriA, if positioned at the back of the replisome, would pull the template for lagging strand synthesis in the opposite direction, enlarging the loop. Priming and elongation of Okazaki fragments can take place in this loop.
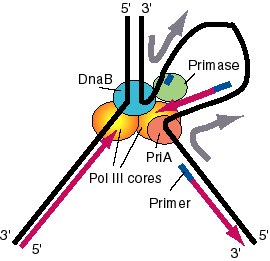
Figure 5.26. A model for the activities of helicases DnaB and PriA at replication fork. In this model, the replication machinery in a large replisome (polymerase and primosome) is postulated to be stationary, with the DNA strands moving through it like fabric through a sewing machine. After unwinding the duplex DNA at the replication fork, the DnaB helicase can also track along single stranded DNA in a 5' to 3' direction. If the helicase is stationary, then the template strand for lagging strand synthesis moves in the direction of the upper gray arrow in the diagram, away from the replication fork. This template strand is also bound to the PriA helicase at the "back" end of the replisome. The 3' to 5' tracking activity of PriA will also pull the template strand, now in the direction of the bottom gray arrow. The result of the DnaB and PriA tracking activities is to pull the template for lagging strand synthesis into a loop, which is tethered to the replisome at both ends. Primase is also in the replisome, and can synthesize primers along this strand at appropriate intervals (thick blue lines), and one of the core polymerases of the DNA polymerase III holoenzyme can synthesize an Okazaki fragment. Newly synthesized DNA is a thick violet line, and parental DNA strands are thick black lines.
Eukaryotic replication proteins have functions analogous to those
found in bacteria.
DNA replication has been studied from a wide variety of species. For our purposes, we will focus on common themes of the mechanisms of replication found both in prokaryotes and in eukaryotes. This section will examine eukaryotic DNA polymerases and accessory proteins, emphasizing properties that are common to those seen in bacterial enzymes.
Five
DNA polymerases, called a, d, b,
e, and g,
have been isolated from
eukaryotic cells. Following the paradigm established for studying replication
in bacteria, researchers have sought to determine which proteins are involved
in a particular function using both genetic analysis and biochemical
characterization. Although no genetic screens for DNA replicating functions can
be done in mammals, a substantial amount has been learned by studying
replication in vitro of DNA containing
viral origins of replication, such as those found in simian virus 40 (SV40) or
bovine papilloma virus. These mammalian viruses have small chromosomes (about 5
to 7 kb), and they can be replicated completely in cell-free systems. Use of cell-free systems that are competent
for replication has allowed a detailed analysis of proteins required for this
process. The ability to interfere with the activity of designated proteins in a
cell-free system, e.g. by adding antibodies that inactivate them or inhibitors
to block their activity, provides the means to test whether the that protein is
required for DNA replication. In effect, this interference with a protein in vitro mimics the information gleaned from the
phenotypes of loss-of-function mutations in the genes that encode the protein
of interest. Also, purified proteins can be combined to reconstitute the
activities needed for complete synthesis of the viral DNA template. Success in
such a reconstitution indicates that the major components have been identified.
Furthermore, proteins homologous to those identified in mammalian cells have
been found in yeast, and mutation of those genes provides additional
information about the biological function of the enzymes. Results of these
types of studies are presented in this section.
The chief polymerase for replication of nuclear DNA is DNA polymerase d (Figure 5.27) . It is required for both leading strand and lagging strand synthesis, at least in reconstituted in vitro replication systems. It has two subunits, a polymerase (125 kDa) and another subunit (48 kDa). It catalyzes DNA synthesis with high fidelity, and it contains the expected 3' to 5' exonuclease activity for proofreading. It has high processivity when associated with an analog of the bacterial sliding clamp (b2), called PCNA.
![]()
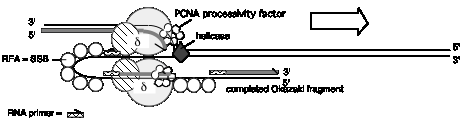
Figure 5.27. Eukaryotic DNA polymerases and replication proteins
at the replication fork. The major replicative DNA polymerase in nuclei is DNA
polymerase d. RFA is the
functional equivalent of bacterial SSB; this single-stranded binding protein
coats the single-stranded DNA. A helicase catalyzes the separation of the two
parental strands. DNA polymerase a (shown as a circle around the new primer)
contains a primase that makes short stretches of RNA and DNA that server as
primers for DNA synthesis.
PCNA, or proliferating cell nuclear antigen, was
initially identified as an antigen that appears only in replicating cells, and
only at certain times of the cell cycle (such as S phase). This trimeric
protein has a ring structure similar to that of the b2
sliding clamp of E. coli DNA polymerase
III (Figure 5.28), despite the absence of significant sequence similarity in
the proteins. Binding of PCNA confers high processivity onto polymerase d.
Thus PCNA is both structurally and functionally analogous to the E.
coli b subunit.
Each subunit of the trimeric PCNA folds into two domains, for a total of six
domains in the ring. Each subunit of the dimeric E. coli b subunit folds into three
domains, again making six domains in the ring. Thus the sliding clamp has a
very similar structure in both bacteria and mammals.

Figure 5.28. Similar structures of processivity factors for DNA
replication. The mammalian protein, PCNA (top), is a trimer, each monomer of which has two similar domains. T he trimer
forms a circle that surrounds DNA, hence serving as a sliding clamp. The
b subunit of DNA polymerase III from E. coli is a dimer (bottom), each
monomer of which has three similar domains. These domains have a very similar
structure to those of PCNA, despite having only limited sequence similarity.
Thus functionally analogous sliding clamps
in eukaryotes and prokaryotes have similar structures.
The template-primer junctions are
recognized by the multisubunit replication factor C, or RFC. Like the g
complex in E. coli, this enzyme is an
ATPase, and it helps to load on the processivity factor PCNA. Thus RFC is
carrying out a similar function to the bacterial g-complex.
One of the first eukaryotic polymerases to be isolated was DNA polymerase a, which is now recognized as a catalyst of primer synthesis. This enzyme contains four polypeptide subunits, one with a polymerase activity (170 kDa), two that comprise a primase activity (50 and 60 kDa), and another subunit of (currently) undetermined function (70 kDa). DNA polymerase a has low processivity but high fidelity. This high fidelity is surprising because no 3' to 5' exonuclease is associated with the enzyme. Polymerase a, possibly with additional primases, catalyzes the synthesis of short segments of DNA and RNA that serve as primers for the replicative polymerases.
DNA polymerase e is related to polymerase d, and it may play a role in lagging strand synthesis. It is also dependent on PCNA, in vivo. However, no requirement has been identified for it in viral replication systems in vitro.
The compound aphidicolin will block the growth of mammalian cells. It
does this by preventing DNA replication, and the targets of this drug are DNA
polymerases a and d (as well as e).
The fact that inhibition of these DNA polymerases with aphidicolin also stops
DNA replication in mammalian cells argues that indeed, a and d are responsible
for replication of nuclear DNA in eukaryotic cells. This conclusion is strongly
supported by the phenotype of conditional loss-of-function mutations in the
genes encoding the homologs to these polymerases in yeast. Such mutants do not
grow at the restrictive temperature, indicating that d and a are the
replicative polymerases. The biochemical evidence implicates polymerase a in primer formation, and d appears to be the major polymerases used to
synthesize the new strands of DNA.
Table 5.4. Analogous components of the replication machinery in E. coli and eukaryotic cells.
|
Function |
Bacterial (E. coli) |
Number of subunits |
Eukaryotic replication (SV40) |
Number of subunits |
|
Leading and lagging strand synthesis |
asymmetric dimer, E. coli polymerase III |
10 (3 in core) |
polymerase d |
2 |
|
Sliding clamp |
b subunit |
2 |
PCNA |
3 |
|
Clamp loader |
g-complex |
6 |
RFC |
multiple |
|
Primase |
DnaG |
1 |
Polymerase a |
4 |
|
Helicase |
DnaB |
6 |
T-antigen (SV40) |
6 |
|
Bind single-stranded DNA |
SSB |
1 |
RFA |
3 |
|
Swivel |
Gyrase |
4, A2B2 |
Topo I or Topo II |
1 2 (homodimer) |
The parallels between bacterial and
eukaryotic DNA replication are striking. The overall strategy of synthesis is
similar, and analogous proteins carry out similar functions, as listed in Table
5.4. It is difficult to determine whether the proteins carrying out similar functions
are actually homologous proteins, i.e. encoded by genes descended from the same
gene in the last common ancestor. The protein sequence identities are marginal,
and frequently the analogous proteins have different numbers of subunits. These
differences complicate the analysis considerably, because different subunits in
bacteria or mammals may have similar functions. However, the functional
similarities are convincing.
Several other DNA polymerases have been isolated from eukaryotic cells. DNA polymerase b and e are involved in repair of nuclear DNA. DNA polymerase b is a single polypeptide of 36 kDa, and has no 3' to 5' exonuclease. DNA polymerase g replicates mitochondrial DNA.
Reverse transcriptase is frequently referred to as an RNA-dependent DNA
polymerase because it can use RNA as a template, but in fact it can use either
RNA or DNA as a template. It is encoded by retroviruses, and hence it is
present in cells infected with a retrovirus. This enzyme has widespread use in
the laboratory for making complementary copies of RNA, called cDNA. Active
copies of LINE1 repetitive elements (in mammals) or Ty1 repeats (in yeast),
also encode reverse transcriptase. Thus in cells where these retrotransposable
elements are being transcribed, active reverse transcriptase is also present.
Reverse transcriptase also has an RNase H activity, which will digest away RNA
from an RNA-DNA duplex.
In contrast to the other DNA polymerases
discussed in this chapter, terminal deoxynucleotidyl transferase does not require a template. It adds dNTPs
(as dNMP) to the 3' end of DNA, using that 3' hydroxyl as a primer. It is found
in differentiating lymphocytes, and appears to be used physiologically to
introduce somatic mutations into immunoglobulin genes. In the laboratory, it is
used to add "homopolymer tails" to the ends of DNA molecules by
incubating a linear DNA with one particular dNTP and terminal deoxynucleotidyl
transferase.
As will be discussed in more detail in the
next chapter, the ends of linear chromosomes (telomeres) must be expanded at
each replication or they will eventually become shortened. The enzyme telomerase catalyzes the addition of many tandem
copies of a simple sequence to the ends of the chromosomes. The template for
this reaction is an RNA that is a component of the enzyme. Thus telomerase is a
reverse transcriptase that only makes copies of the template that it carries,
using the 3' end of a chromosomal DNA strand as the primer.
Further
readings
Advanced
text:
A. Kornberg and T. Baker (1992) DNA Replication, 2nd Edition, W.H. Freeman and Company, New York.
Classic
papers:
A. Kornberg, I. R. Lerman, M. J. Bessman, and E. S. Simms (1956) "Enzymic synthesis of deoxyribonucleic acid" Biochimica et Biophysica Acta 21:197-198.
M. Meselson and F. W. Stahl (1958) "The replication of DNA in Escherichia coli." Proceedings of the National Academy of Sciences, USA 44:671-682.
R. Okazaki, T. Okazaki, K. Sakabe, K. Sugimoto, and A. Sugino (1968) “Mechanism of DNA Chain Growth, I. Possible Discontinuity and Unusual Secondary Structure of Newly Synthesized Chains” Proceedings of the National Academy of Sciences, USA 59: 598-605.
P. De Lucia and J. Cairns (1969) Isolation of an E. coli strain with a mutation affecting DNA polymerase. Nature 224:1164-1166.
J. Gross and M. Gross (1969) Genetic analysis of an E. coli strain with a mutation affecting DNA polymerase. Nature 224:1166-1168
Recent
reviews:
R. Sousa (1996) Trends in Biochemical Sciences 21:186-190. Similarities in structure among DNA polymerases
Herendeen and Kelly (1996) Cell 84:5-8. Subunits and mechanism of DNA polymerase III.
Questions and Problems
DNA Replication I
Chapter 5
Question 5.8 Imagine you are investigating the replication of a bacterial species called B. mulligan. The bacteria is grown for several generations in medium containing a heavy density label, [15N] NH4Cl. The bacteria are then shifted to medium containing normal density [14N] NH4Cl. DNA is extracted after each generation and analyzed on a CsCl gradient. From the results shown below, what is the mode of replication in B. mulligan? Explain your conclusion.
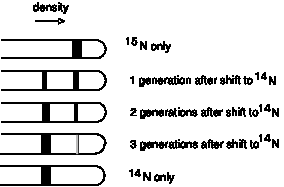
Question 5.9 How many turns must be unwound during replication of the E. coli chromosome? The chromosome contains 4.64 x 106 base pairs.
Question 5.10 Which of the following comments about Okazaki fragments are true or false? Okazaki fragments:
a) are short segments of newly synthesized DNA.
b) are formed by synthesis on the leading strand of DNA.
c) have a short stretch of RNA, or a mixture of ribonucleotides and deoxyribonucleotides, at their 5' end.
d) account for overall synthesis of one DNA strand in a 3' to 5' direction.
Question 5.11.
The following experimental results are from A. Sugino and R. Okazaki (1972)
“Mechanisms of DNA Chain Growth VII. Direction and rate of growth of T4 nascent
short DNA chains” J. Mol. Biol. 64: 61-85.
a. E. coli cells were infected with bacteriophage T4
and then chilled to 4oC to slow the rate of replication. Replicating
DNA in the infected cells was pulse-labeled with [3H]-thymidine (a) or [3H]-thymine
(b) for 5 sec (black-filled circles), 30 sec (open circles with vertical line),
60 sec (open circles with dot) or 300 sec (open circles). The pulse labeling
was stopped with potassium cyanide and ice, and the DNA was extracted,
denatured in NaOH, and separated on an alkaline sucrose gradient. Fractions
from the gradient were collected and assayed for the amount of 3H in
the DNA (as material that bound to a filter after washing in (a) and as
acid-insoluble material in (b)). The sedimentation value in Svedbergs (S) is
given along the x-axis; faster sedimenting material is toward the right. What
do these data tell you about the sizes of nascent (newly synthesized) DNA at
the various pulse labeling times?
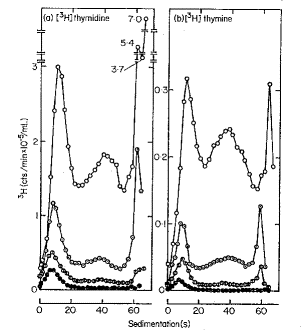
(b) Sugino and Okazaki used a method to
break the isolated short nascent chains (completed Okazaki fragments) randomly
and recover only the oligonucleotides from the 5í ends. They found that at very
short labeling times (e.g. 5 sec) the [3H] thymidine was not at the
5’ ends of the DNA (hence it was internal and at the 3’ ends). After longer
labeling times, the [3H] thymidine was found in the oligonucleotides
at the 5’ end. What do you conclude is the direction of chain growth of the
nascent chains? Explain your logic.
Question 5.12 We have covered two experiments from the Okazaki lab using pulse labeling for increasing times to follow the synthesis of new DNA. How would you design a pulse-chase experiment to monitor not only the initial production of Okazaki fragments, but also their incorporation into larger DNA molecules?
Question 5.13 Which enzymes, substrates, and cofactors are used in common and which ones are distinctive for synthesis of leading strands and lagging strands of DNA at the replication fork of E. coli?
Question 5.14 Which subunit or complex within E. coli DNA polymerase III holoenzyme has each the following functions?
a) Catalyzes 5'to 3' polymerization of new DNA.
b) Has the proofreading function (3' to 5' exonuclease).
c) Dimerizes the two catalytic cores.
d) Forms the clamp that is thought to account for its high processivity.
e) Loads and unloads the sliding clamp.
Question 5.15 What are the components of the multiprotein complex known as the primosome in E. coli? What does it do? In what direction does it travel?
Question 5.16 Which eukaryotic nuclear DNA polymerase(s) is (are) thought to account for leading strand and lagging strand synthesis?