B M B 400, Part
Three
Gene Expression
and Protein Synthesis
Chapter 12 RNA
PROCESSING
A. Types of RNA processing
1. RNA processing refers to any covalent modification to the RNA that occurs after transcription. This includes specific cleavage, addition of nucleotides, methylation or other modification of the nucleotides, and removal of introns by splicing.
2. Overview
|
RNA |
Precursor |
Modification |
Addition |
Cleavage |
Splicing |
|
mRNA |
pre‑mRNA (hnRNA) |
methylation on 2'‑OH of ribose |
5' cap 3' poly A |
cut at site for poly A; excise viral mRNA |
remove introns |
|
rRNA |
pre‑rRNA |
methylation on 2'OH of ribose |
no |
excise products fr. precursor |
remove introns |
|
tRNA |
pre‑tRNA |
extensive and varied |
CCA to 3' end |
yes |
remove introns |
|
snRNAs |
? |
? |
5' cap |
? |
? |
B. Cutting and trimming RNA
1. pre‑rRNA
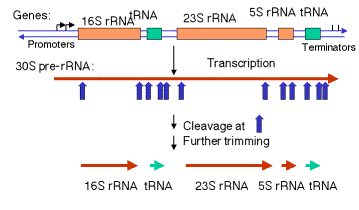
a. In E. coli, the rrn operon is transcribed into a 30S precursor RNA, containing 3 rRNAs and 2 tRNAs.
Figure
3.3.1. Excision of rRNAs and tRNAs from 30S precursor RNA
(1) The segment containing 16S rRNA (small ribosomal subunit) and the one containing 23S rRNA (large ribosomal subunit) are flanked by inverted repeats that form stem structure in the RNA.
(2) The stems are cleaved by RNase III. There is no apparent single sequence at which RNase III cleaves ‑ perhaps it recognizes a particular stem structure. This plus subsequent cleavage events (by an activity called M16) generates the mature 16S and 23S rRNAs. The rRNAs are also methylated.
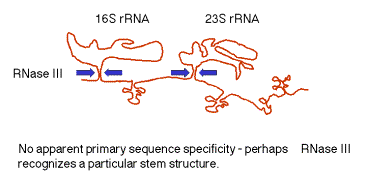
Figure
3.3.2. RNase III cuts in the stems
of stem-loops in RNA
(3) tRNA is liberated by RNases P and F.
(4) 5S rRNA is liberated by RNases E and M5
b. In eukaryotes:
(1) The initial precursor is 47S and contains ETS1, 18S rRNA, ITS1, 5.8S rRNA, ITS2, and 28S rRNA, where ETS = extragenic transcribed spacer and ITS = intragenic transcribed spacer.
(2) Specific cleavage events followed by methylations generate the mature products. Also, some rRNA genes in some species have introns that must be spliced out.
2. pre-tRNA
in E. coli
a. Sequence specific cleavage by RNases P, F, D
(1) RNase P is an endonuclease that cleaves the precursor to generate the 5' end of the mature tRNA.
(2) RNase F is an endonuclease that cleaves the precursor 3 nucleotides past the 3' end of the mature tRNA.
(3) RNase D is an exonuclease that trims in a 3' to 5' direction to generate the 3' end or the mature tRNA.
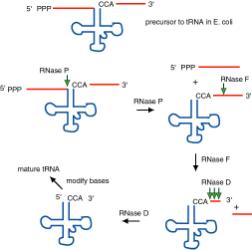
Figure 3.3.3. The ends of tRNA in E. coli are produced by the action of three nucleases that cleave the precursor to tRNA. A schematic of the pre-tRNA is shown at the top, with RNA extending from the 5’ and 3’ ends of the RNA that will become the mature tRNA (shown as a cloverleaf). The site of cleavage is indicated by the short vertical arrows above the lines denoting RNA, and they are labeled with the name of the enzyme cutting at that site. The enzymes catalyzing each reaction are listed above or adjacent to the reaction arrows.
b. The catalytic activity of RNase P is in the RNA component
(1) RNAse P is composed of a 375 nt RNA and a 20 kDa protein.
(2) The catalytic activity is in the RNA. The protein is thought to aid in the reaction, but is not required for catalysis. All enzymes are not proteins!
(3) This was one of the first instances discovered of catalytic RNA, and Sidney Altman shared the Nobel Prize for this.
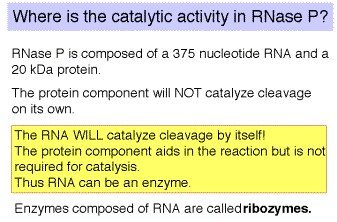
Fig.
3.3.4. RNase P
c. The enzyme tRNA nucleotidyl transferase adds CCA to the 3' ends of pre-tRNAs.
(1) Virtually all tRNAs end in CCA, forms the amino acceptor stem.
(2) For most prokaryotic tRNA genes, the CCA is encoded at the 3' end of the gene.
(3) No known eukaryotic tRNA gene encodes the CCA, but rather it is added posttranscriptionally by the enzyme tRNA nucleotidyl transferase. This enzyme is present in a wide variety of organisms, including bacteria, in the latter case presumably to add CCA to damaged tRNAs.
C. Modifications at the 5' and 3' ends of
mRNA
As discussed previously, eukaryotic mRNAs are capped at their 5' end and polyadenylated at their 3' end. In vitro assays for these reactions have been developed, and several of the enzymatic activities have been identified. These will be reviewed in this section. Polyadenylation is not limited to eukaryotes. Several mRNAs in E. coli are polyadenylated as well. This is a fairly new area of study.
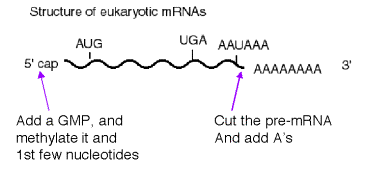
Fig. 3.3.5. mRNAs
can be modified on the 5’ and 3’ ends.
1. Modification at the 5' end: cap structure
a. The "cap" is a methylated 5'‑GMP that is linked via its 5' phosphate to the b‑phosphoryl of the initiating nucleotide (usually A). See Fig. 3.3.6.
b. Capping occurs shortly after transcription has begun.
c. It occurs in a series of enzymatic steps (Fig. 3.3.7).
(1) Remove the g‑phosphoryl of the initiating nucleotide (RNA triphosphatase)
(2) Link a GMP to the b‑phosphoryl of the initiating nucleotide (mRNA guanylyl transferase). The GMP is derived from GTP, and is linked by its 5' phosphate to the 5' diphosphate of the initiating nucleotide. Pyrophosphate is released.
(3) The N‑7 of the cap GMP is methylated (methyl transferase), donor is S‑adenosyl methionine.
(4) Subsequent methylations occur on the 2' OH of the first two nucleotides of the mRNA.
d. Capping has been implicated in having a role in efficiency of translation and in mRNA stability.
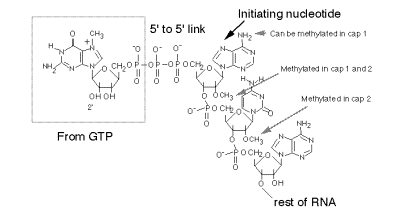
Fig.
3.3.6. Structure of the 5’ cap on
eukaryotic mRNAs.
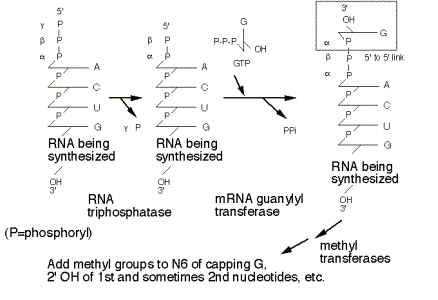
Figure
3.3.7. Stepwise synthesis of the
5’ cap.
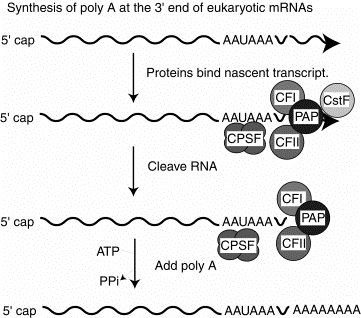
2. Several proteins are required for cleavage and polyadenylation at the 3' end.
CPSF is a tetrameric specificity factor; it recognizes and binds to the AAUAAA polyadenylation signal.
CFI and CFII are cleavage factors.
PAP is the polyA polymerase.
CFI, CFII and PAP form a complex that binds to the nascent RNA at the cleavage site, directed by the CPSF specificity factor.
CstF is an additional protein implicated in this process in vitro, but its precise function is currently unknown.
Fig. 3.3.8
D. Multiple mechanisms are used for splicing different types of introns.
1. Different
types of introns
a. pre‑tRNA
b. group I, group II: Introns in fungal mitochondrial genes and in plastid (chloroplast) genes have been grouped into 2 different groups based on different consensus sequences found in the introns. As we will see below, the group II introns have a mechanism for splicing that is similar to that of pre‑mRNA.
c. pre‑mRNA
In all cases, splicing will remove the introns and join the exons to give the mature RNA.
Table. Features of splicing for different types of introns
|
Class |
Distribution |
Sequence |
Distinguishing feature |
Mechanism |
|
pre-tRNA |
yeast to mammals |
very short (10-20 nucleotides) |
requires ATP |
cut, kinase, ligase |
|
group I |
fungal mitochondria, plastids, pre-rRNA in Tetrahymena |
characteristic consensus |
self-splicing, G nucleot(s)ide to initiate |
phosphoester transfer |
|
group II |
fungal mitochondria, plastids |
characteristic consensus |
can self-splice, internal A nucleotide to initiate |
phosphoester transfer |
|
pre-mRNA |
yeast to mammals |
5' GU...AG 3' |
spliceosome (ATP for assembly), internal A nucleotide to initiate |
phosphoester transfer |
2. Splicing
of pre‑tRNAs
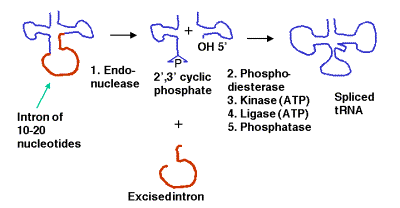
a. Some precursor tRNAs contain short introns (only 10 to 20 nucleotides) with no apparent consensus sequences.
b. These short introns are removed in a series of steps catalyzed by enzymes. The enzymes include an endonuclease, a kinase and a ligase. Because the endonuclease generates a 2’, 3’ cyclic phosphodiester product, an additional phosphodiesterase is needed to open the cyclic phosphodiester to provide the 3’ hydroxyl for the ligase reaction. In addition, the 2’-phosphate (product of the phosphodiesterase) must be removed by a phosphatase.
c. This process uses 2 ATPs for every splicing event.
E. Self‑splicing by group I introns (pre‑rRNA of Tetrahymena)
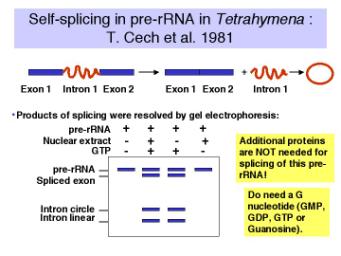
1. Discovery of self-splicing
An in vitro reaction was established to examine the removal of an intron from the precursor to rRNA in Tetrahymena. Suprisingly, it was discovered that the splicing of the pre-RNA occurred in the absence of any added protein!
Figure 3.3.10. Discovery of self-splicing in T. Cech’s lab, 1981
Further investigation revealed the following.
a. The reaction requires a guanine nucleotide or nucleoside with a 3'‑OH, plus mono‑ and divalent cations. GTP, GDP, GMP or guanosine will work to initiate splicing.
b. There is no requirement for protein or high energy bond cleavage.
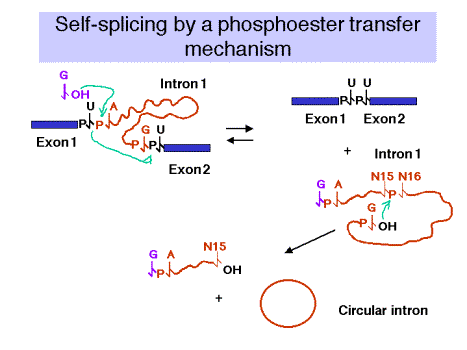
2. Self‑splicing occurs by a phosphoester transfer mechanism (Fig. 3.3.11)
(1) The 3'‑OH of the guanine nucleotide is the nucleophile that attacks and joins to the 5' phosphate of the first nucleotide of the intron.
(2) This leaves the 3'‑OH of the last nucleotide of the upstream exon available to attack and join the 5' phosphate of the first nucleotide of the downstream exon.
(3) These two phosphoester transfers result in a joining of the two exons and excision of the intron (with the initiating G nucleotide attached to the 5' end.)
(4) The excised intron is then circularized by attack of the 3'‑OH of the last nucleotide of the intron on the phosphate between the 15th and 16th nucleotides of the introns. Further degradation effectively removes the intron from the reaction and helps prevent the reverse reaction from occurring. Note that the phosphoester transfers are readily reversible unless the products (excised intron) are removed.
(5) There is no increase or decrease in the number of phosphoester bonds during this splicing.
Figure 3.3.11.
3. The intron is the catalyst for splicing in this system.
a. RNA involvement in self‑splicing is stoichiometric, but the excised intron does have a catalytic activity in vitro.
After a series of intramolecular cyclization and cleavage reactions, the linear excised intron lacking 19 nucleotides (called L-19 IVS) can be used catalytically to add and remove nucleotides to an artificial substrate. For instance, C5, which is complementary to the internal guide sequences of the intron, can be converted to C4 + C6 and other products (Fig. 3.3.12).
b. The 3‑D structure of the folded RNA is responsible for the specificity and efficiency of the reaction (analogous to the general ideas about proteins with enzymatic activity). The specificity of splicing is caused, at least in part, by base‑pairing between the 3' end of the upstream exon and a region in the intron called the internal guide sequence. The initiating G nt also binds to a specific site in the RNA close to the 5' splice site. Thus two sites in the pre-rRNA intron are used sequentially in splicing (Fig. 3.3.13 A and 3.3.13.B.).
Figure 3.3.12.

Fig. 3.3.13.A.
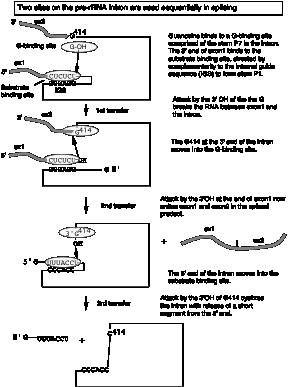
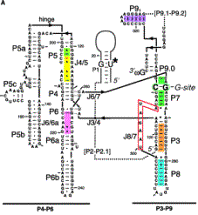
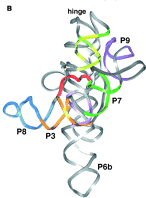
Fig. 3.3.13.B. The catalytic domain of the group I intron from Tetrahymena pre-rRNA, shown in the RNA secondary structure view (left panel) and in a view of the tertiary structure (right panel).
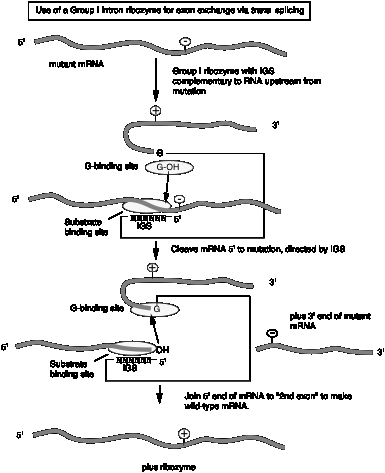
c. The internal guide sequence (IGS) is not not required for catalysis but does confer specificity. Thus one can design RNAs for exon exchange in cells. This potential avenue for therapy for genetic disorders is called "exon replacement therapy.
Fig.
3.3.14.
F. RNAs can function as enzymes
Examples include the following:
RNase P
Group I introns (includes intron of pre‑rRNA in Tetrahymena)
Group II introns
RNA: peptide bond formation
Hammerhead
ribozymes
Viroids and virusoids have a self-cleaving activity that localized to a 58 nucleotide structure, illustrated in Fig. 3.3.15.
The mechanism differs in some respects from the phosphoester transfer. A divalent metal hydroxide binds in the active site, and abstracts a proton from the 2' OH of the nucleotide at the cleavage site. This now serves as a nucleophile to attack the 3' phosphate and cleave the phosphodiester bond, generating a 2',3' cyclic phosphate and a 5' OH on the ends of the cleaved RNA.
Fig. 3.3.15.
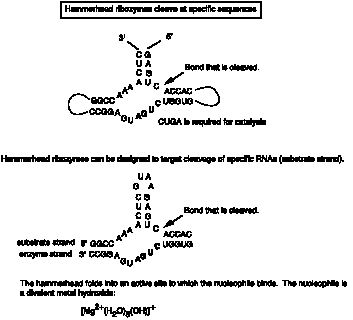
One application currently being explored is the use of designed hammerheads to cleave a particular mRNA, thereby turning off expression of a particular gene. If over-expression or ectopic expression of a defined gene were the cause of some pathology (e.g. some form of cancer), then reducing its expression could have therapeutic value.
Other RNAs possibly involved in catalysis, such as the snRNAs involved in splicing pre-mRNA (discussed in the next section).
Even though RNAs can be sufficient for catalysis, sometimes they are assisted by proteins to improve efficiency. For instance, group I introns are capable of splicing introns by themselves in a cell-free reaction. However, some are not very efficient in this process, and in the cell they are assisted by proteins that themselves are not catalytic but they enhance the reaction. Examples are maturases, which are proteins that assist in the splicing of some group I introns found in yeast mitochondria.
G. Splicing
of introns in pre‑mRNAs
1. The sequence at the 5' and 3' ends of introns in pre-mRNAs is very highly conserved.
Thus one can derive a consensus sequence for splice junctions.
(1) 5' exon...AG'GURAGU.................YYYYYYYYYYNCAG'G....exon
The GU is the 5' splice site (sometimes called the donor splice site), and the AG is the 3' splice site (or accepter splice site).
(2) GU is invariant at the 5' splice site, and AG is (almost) invariant at the 3' splice site for the most prevalent class of introns in pre-mRNA.
(3) Effects of mutations at the splice junctions demonstrate their importance in the splicing mechanism.
Mutation of the GT at the donor site in DNA to an AT prevents splicing (this was seen in a mutation of the b‑globin gene that caused b0 thalassemia.) A different mutation of the b‑globin gene that generated a new splice site caused an aberrant RNA to be made, resulting in low levels of b‑globin being produced (b+ thalassemia).
2. The intron is excised as a lariat.
a. The 2'‑OH of an A at the "branch" point forms a phosphoester with the first G of the intron to initiate splicing.
b. Splicing occurs by a series of phosphoester transfers (also called trans‑esterifications). After the 2'-OH of the A at the branch has joined to the initial G of the intron, the 3'‑OH of the upstream exon is available to react with the first nucleotide of the downstream exon, thereby joining the two exons via the phosphoester transfer mechanism.
c. Intron lariat is the equivalent of a "circular" intermediate.
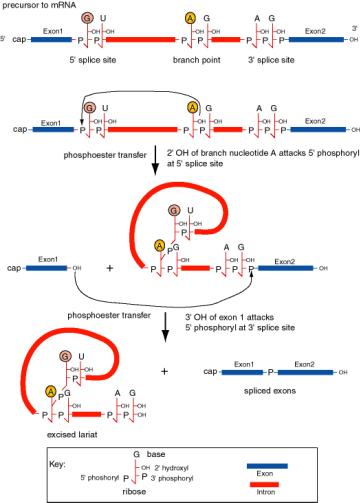
Figure 3.3.16. Splicing of precursor to mRNA excises the intron as a lariat structure. The chemical reactions are two phosphoester transfers. The first transfer is initiated by the 2’ hydroxyl of the adenine ribonucleoside at the branch point, which attacks the 5’ phosphoryl of the 5’ splice site. This generates a 3’ hydroxyl at exon 1 and joins the A at the branch point to the U at the 5’ splice site, producing a lariat in the intron. The second transfer is initiated by the attack of the newly exposed 3’ hydroxyl of exon 1 on the 5’ phosphoryl of exon 2. The latter reaction joins the two exons and releases the intron as a lariat.
d. The sequence at the branch point is only moderately conserved in most species; examination of many branch points gives the consensus YNYYRAG. It lies 18 to 40 nucleotides upstream of the 3' splice site.
3. Small nuclear ribonucleoproteins (or snRNPs) form the functional splicesome on pre‑mRNA and catalyze splicing.
a. "U" RNAs and associated proteins
Small nuclear RNAs (snRNAs) are about 100 to 300 nts long and can be as abundant as 105 to 106 molecules per cell. They are named U followed by an integer. The major ones involved in splicing are U1, U2, U4/U6, and U5 snRNAs. They are conserved from yeast to human.
The snRNAs are associated with
proteins to form small nuclear ribonucleoprotein particles, or snRNPs. The snRNPs are named for the snRNAs they contain,
hence the major ones involved in splicing are U1, U2, U4/U6, U5 snRNPs.
One class of proteins common to many snRNPs are the
Sm proteins.
There are 7 Sm proteins, called B/B’, D1, D2, D3, E, F, G. Each Sm protein has
similar 3-D structure, consisting of an alpha helix followed by 5 beta strands.
The Sm proteins interact via the beta strands, and may form circle around RNA.
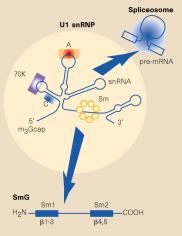
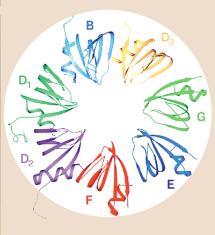
Fig. 3.3.16.1. In the U1 snRNP (left panel),
the Sm protein SmG is thought to interact with other Sm proteins to form a ring
around the U1snRNA at a motif just before the 3’ stem-loop. Other proteins (A,
C, 70K) interact with other parts of the U1 RNA, which is then asssembled into
a large spliceosome (see Fig. 3.3.17). The right panel shows interactions of
the Sm proteins through their beta-strands to make a ring with an inner portion
large enough to encircle an RNA molecule. From Angus I. Lamond (1999) Nature
397, 655 - 656 “RNA splicing: Running rings around RNA.”
A particular sequence common to many snRNAs is recognized by the Sm proteins, and is called the “Sm RNA motif”.
b. Use of antibodies from patients with SLE
Several of the common snRNPs are recognized by the autoimmune serum called anti‑Sm, initially generated by patients with the autoimmune disease Systemic Lupus Erythematosis. One of the critical early experiments showing the importance of snRNPs in splicing was the demonstration that anti-Sm antisera is a potent inhibitor of in vitro splicing reactions. Thus the targets of the antisera, i.e. Sm proteins in snRNPs, are needed for splicing.
c. The snRNPs assemble onto the pre-mRNA to make a large protein-RNA complex called a spliceosome (Fig. 3.3.17). Catalysis of splicing occurs within the spliceosome. Recent studies support the hypothesis that the snRNA components of the spliceosome actually catalyze splicing, providing another example of ribozymes.
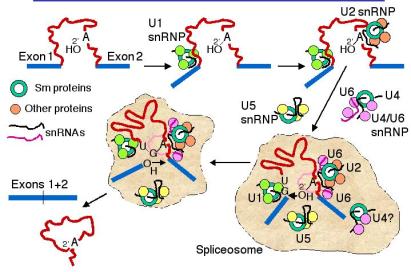
Figure 3.3.17. Spliceosome assembly and catalysis
d. U1 snRNP: Binds to the 5' splice site, and U1 RNA forms a base‑paired structure with the 5' splice site.
e. U2 snRNP: Binds to the branch point and forms a short RNA-RNA duplex. This step requires an auxiliary factor (U2AF) and ATP hydrolysis, and commits the pre-mRNA to the splicing pathway.
f. U5 snRNP plus the U4, U6 snRNP now bind to assemble the functional spliceosome. Evidence indicates that U4 snRNP dissociates from the U6 snRNP in the spliceosome. This then allows U6 RNA to form new base-paired structures with the U2 RNA and the pre-mRNA that catalyze the transesterification reaction (phosphoester transfers). One model is that U6 RNA pairs with the 5' splice site and with U2 RNA (which itself is paired to the branch point), thus bringing the branch point A close to the 5' splice site. U5 RNA may serve to hold close together the ends of the exons to be joined.
4. Trans‑splicing
All of the splicing we have discussed so far is between exons on the same RNA molecule, but in some cases exons can be spliced to other RNAs. This is very common in trypanosomes, in which a spliced leader sequence is found at the 5' ends of almost all mRNAs. A few examples of trans splicing have been described in mammalian cells.
H. Splicing
of group II introns
1. Similar mechanism as that for nuclear pre‑mRNA splicing.
2. Can occur by self‑splicing, albeit under rather artificial conditions.
3. Reaction can be reversible (as can splicing of group I introns), leading to the idea that these introns can be transposable elements.
4. The group II self‑splicing may be the evolutionary ancestor to nuclear pre‑mRNA splicing.
I. Mechanistic
similarties for splicing group I, group II and pre‑mRNA introns
1. All involve transesterification = phosphoester transfers. No high energy bonds are utilized in the splicing process; the arrangement of phosphodiester bonds is reorganized, and as a result exons are joined together.
2. The initiating nucleophile is the 3' OH of a guanine nucleotide for Group I introns, whereas for Group II introns and introns in pre‑mRNA, it is the 2' OH of an internal adenine nucleotide in the intron.
3. In all cases, particular secondary structures in the RNAs are utilized to bring together the reactive components (e.g. ends of exons and introns). These secondary structures may be intramolecular in the case of self‑splicing Group I and Group II introns, or they may be intermolecular in the case of pre‑mRNA and the snRNAs, e.g. those in the U1, U2, perhaps U6 snRNPs.
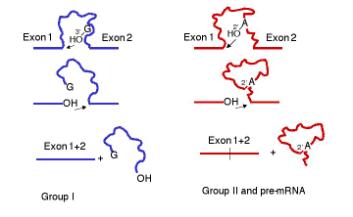
Figure 3.3.18. Common features of the mechanism of splicing in Group I introns and in Group II introns plus introns in precursor to mRNA.
J. Alternative splicing
1. General comments
a. For many genes, all the introns in the mRNA are spliced out in a unique manner, resulting in one mRNA per gene. But there is a growing number of examples of other genes in which certain exons are included or excluded from the final mature mRNA, a process called alternative splicing.
b. Some exons may be included in some tissues and not others, or may be sex‑specific, indicating some regulation over the selection of splice sites.
c. Alternative splicing of pre‑mRNA means that a single gene may encode more than one protein product.
2. Specific example: Sex determination in Drosophila melanogaster
a. The X to autosome ratio (X:A ratio) in the zygote will determine which of two different developmental pathways along which the fly will develop. If the X:A ratio is high (e.g. the female is XX and the X:A ratio is 1.0), the fly will utilize the female pathway; if the ratio is low (e.g. 0.5 since the male is XY), it will develop as a male.
The X:A ratio is determined by "counting" certain genes (or their expression) on the X chromosome (e.g. sisterless a, sisterless b, and runt) for the numerator and counting other genes (such as deadpan) for the denominator. All of the products of these genes are homologous to various calsses of transcription factors, consistent with at least part of the regulation of sex determination being transcriptional. However, as discussed below, alternative splicing plays a key role as well, at least in Drosophila.
b. The pathways have at least 4 steps that were defined genetically by mutations that caused, e.g. genetically female flies (high X:A) to develop as males. In each case, the same gene encodes both male and female‑specific mRNAs (and proteins), but the sex‑specific mRNAs (and proteins) differ as a result of alternative splicing.
c. In all cases, the default state is male development, and some new activity has to be present to establish and maintain the female pathway.
(1). The target of the X:A signal is the Sex-lethal gene (Sxl), which serves as a master switch gene. In early development, an X:A ratio of 1 in females leads to the activiation of an embryo-specific promoter of the Sxl gene, whereas Sxl is not transcribed in male embryos. Later in development, Sxl is transcribed in both sexes. Now the high X:A ratio leads to the skipping of an exon in the splicing of pre‑mRNA from the Sex‑lethal gene. This produces a functional Sxl protein in females. In males (default pathway), the mRNA has an early termination codon, and no functional Sxl protein is made.
Figure 3.3.19.
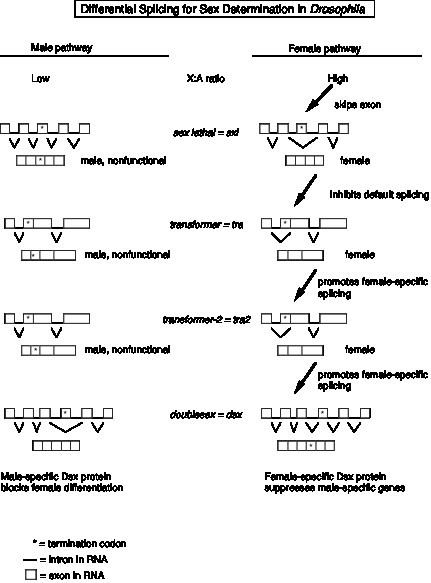
(2) A functional Sxl protein inhibits the default splicing of pre‑mRNA from the transformer gene, to generate a functional Tra protein in female embryos. In the female‑specific splicing of tra pre‑RNA, a 5' splice site (common to both male and female splicing) is connected to an alternative 3' splice site, thereby removing a termination codon and allowing function Tra protein to be made (Fig. 3.3.15).
(3) The Tra protein promotes female‑specific splicing of pre‑mRNA from the tra2 gene, again generating a functional Tra2 protein only in females.
(4) Tra and Tra2 proteins promote female‑specific splicing of pre‑mRNA from the doublesex gene (dsx). In this case, the male‑specific mRNA has skipped an exon (Fig. 3.3.15). Skipping an exon requires an alteration in the splicing pattern at both the 3' splice site and the 5' splice sites surrounding the exon.
(5) The male‑specific Dsx protein blocks female differentiation and leads to male development. The female‑specific Dsx protein represses expression of male genes and leads to female development.
d. Some clues about mechanism
(1) Tra and Tra2 are RNA‑binding proteins related to Splicing Factor 2 (SF2). This latter protein has a domain rich in the dipeptide Arg‑Ser, which defines one type of RNA‑binding domain. SF2 is required for early steps in spliceosome assembly. The related Tra and Tra2 proteins are not required for viability, but they do regulate the specific splicing events for pre‑mRNA from dsx.
(2) Tra2 binds in the female‑specific exon of the dsx transcript, and presumably regulates splice site selection. The binding site for Tra2 within the exon is an example of a splicing enhancer. The mechanisms by which the binding of splicing regulatory proteins (e.g. Tra, Tra2) to splicing enhancers is a very active area of research currently.
(3) Sxl is another RNA‑binding protein that inhibits the default splicing pattern for tra pre‑mRNA.
Figure 3.3.20.
K. RNA editing
1. RNA editing refers to changing the sequence of RNA after transcription, either by adding nucleotides, taking them away, or substituting one for another.
2. The extent of editing is dramatic in some mRNAs, e.g. in the mitochondria of trypansomes and Leishmania.
a. For some mRNAs 55% of the nucleotide sequence is added after transcription! In many of the cases characterized so far, a small number of U's are inserted at many places in the mRNA.
b. Other examples of excising U's and adding C's are known for other mitochondrial genes from other organisms.
3. In at least some cases, the additional nucleotides are added under the direction of guide RNAs that are encoded elsewhere in the mitochondrial genome.
a. A portion of the guide RNA is complementary to the mRNA in the vicinity of the position at which nucleotides will be added (Fig. 3.3.16).
b. The U at the 3' end of the guide RNA initiates a series of phosphoester transfer reactions to insert itself into the mRNA (see bottom of Fig. 3.3.16).
c. More U's at the 3' end of the guide RNA can be added, one at a time.
d. Note the similarity in mechanism between these insertions of nucleotides (editing) and the self‑splicing of Group I intron.
3. For a situation in which one segment of DNA encodes the unedited mRNA and two other segments of DNA encode the guide RNAs required for editing, the "gene" is encoded in three portions, mutations in which would complement in trans! This is a counter‑example to one of our most powerful definitions of a gene.
4. In mammals, two different forms of apolipoprotein B are made, one in the liver and one in the intestine. The intestinal form is much shorter because of an earlier termination codon. Surprisingly, only one gene is found and it must encode both from of ApoB. A specifc enzyme must change one nucleotide of the mRNA for apolipoprotein B (a C in codon 2153, CAA) post‑transcriptionally from a C to a U to generate the termination codon (UAA) found in the intestinal form.
This enzymatic activity is present in a protein with no apparent RNA component, and hence no obvious guide RNA. Thus it appears to operate by a distinctly different mechanism from the editing in protist mitochondria (see. e.g. Greeve, J. et al., 1991, Nucleic Acids Research 19: 3569-3576).
Chapter 12. Questions on RNA processing
12.1 Nucleoside triphosphates labeled with [32P] at the a, b, or g position are useful for monitoring various aspects of transcription. For the specific process listed in a-c, give the position of the label that is appropriate for examining that step.
a) Initiation by E. coli RNA polymerase.
b) Forming the 5' end of eukaryotic mRNA.
c) Elongation by eukaryotic RNA polymerase II.
12.2 (POB) RNA posttranscriptional processing.
Predict the likely effects of a mutation in the sequence (5')AAUAAA in a eukaryotic mRNA transcript.
12.3 A phosphoester transfer mechanism (or transesterification) is observed frequently in splicing and other reactions involving RNA. Are the following statements about these mechanisms true or false?
a) The mechanism requires the cleavage of high-energy bonds from ATP.
b) The initiating nucleophile for splicing of Group I introns (including the intron of pre-rRNA from Tetrahymena) is the 3' hydroxyl of a guanine nucleotide.
c) The initiating nucleophile for splicing of nuclear pre-mRNA is the 2' hydroxyl of an internal adenine nucleotide.
d) The individual reactions in the phosphoester transfers are reversible, but the overall process is essentially irreversible because of circularization (includes lariat formation) of the excised intron.
12.4 What properties are shared by the splicing mechanism of Tetrahymena pre-rRNA and Group II fungal mitochrondrial introns?
12.5 Please answer these questions on splicing of precursors to mRNA.
a) What dinucleotides are almost invariably found at the 5’ and 3’ splice sites of introns?
b) Which splicing component binds at the 5' splice junction?
c) What nucleotides are joined by the branch structure in the intron during splicing?
d) What is ATP used for during splicing of precursors to mRNA?
12.6 (POB) RNA splicing.
What is the minimum number of transesterification reactions needed to splice an intron from an mRNA transcript? Why?
12.7 Match the following statements with the appropriate eukaryotic splicing process listed in parts a-c.
1) A guanine nucleoside or nucleotide initiates a concerted phosphotransfer reaction.
2) The consensus sequences at splice junctions are AG'GUAAGU...YYYAG'G (' is the junction, Y = any pyrimidine).
3) Splicing occurs in two separate steps, cutting to generate a 3'-phosphate followed by an ATP dependent ligation.
4) Splicing requires no protein factors.
5) Splicing requires U1 small nuclear ribonucleoprotein complexes.
a) Splicing of pre-mRNA.
b) Splicing of pre-tRNA in yeast
c) Splicing of pre-rRNA in Tetrahymena
12.8 The enzyme RNase H will cleave any RNA that is in a heteroduplex with DNA. Thus one can cleave a single-stranded RNA in any specific location by first annealing a short oligodeoxyribonucleotide that is complementary to that location and then treating with RNase H.

This approach is useful in determining the structure of splicing intermediates. Let's consider a hypothetical case shown in the figure below. After incubation of radiolabeled precursor RNA (exon1-intron-exon2) with a nuclear extract that is capable of carrying out splicing, the products were analyzed on a denaturing polyacrylamide gel. The results showed that the exons were joined as linear RNA, but the excised intron moved much slower than a linear RNA of the same size, indicative of some non-linear structure. The excised intron was annealed to a short oligodeoxyribonucleotide that is complementary to the region at the 5' splice site (labeled oligo 1 in the figure), treated with RNase H and analyzed on a denaturing polyacrylamide gel. The product ran as a linear RNA with the size of the excised intron (less the length of the RNase H cleavage site). As summarized in the figure, the excised intron was analyzed by annealing (separately) with three other oligodeoxyribonucleotides, followed by RNase H treatment and gel electrophoresis. Use of oligodeoxyribonucleotide number 2 generated a Y-shaped molecule, use of oligodeoxyribonucleotide number 3 generated a V-shaped molecule with one 5' end and 2 3' ends, and use of oligodeoxyribonucleotide number 4 generated a circle and a short linear RNA.
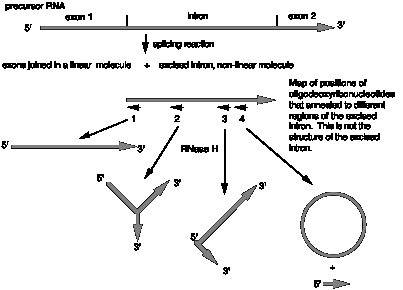
(a) What does the result with oligodeoxyribonucleotide 2 tell you?
(b) What does the result with oligodeoxyribonucleotide 4 tell you?
(c) What does the result with oligodeoxyribonucleotide 1 tell you?
(d) What does the result with oligodeoxyribonucleotide 3 tell you?
(e) What is the structure of the excised intron? Show the locations of the complementary oligos on your drawing.