This
chapter was published in Disorders of Hemoglobin: Genetics, Pathophysiology, Clinical Management in 2001, Cambridge University Press,
Cambridge, UK
Editors: Martin H. Steinberg, Bernard G. Forget,
Douglas R. Higgs, and Ronald L. Nagel
Copyright: Cambridge University Press and Ross
Hardison
Chapter 5:
Organization, evolution and regulation of the globin genes
Original
version completed: June 17, 1998
Updated
May 09, 2000
Published
in 2001.
The
revised version of this book (completed in 2007) leaves out most of the
material in this chapter, and thus I am posting it on the internet to keep the
material available. Please reference the chapter and book if you use this
information.
Author: Ross C. Hardison
Department
of Biochemistry and Molecular Biology, Center for Comparative Genomics and
Bioinformatics, The Pennsylvania State University, 304 Wartik Laboratory,
University Park, PA, 16802 USA
Phone:
814-863-0113
E-mail:
rch8@psu.edu
5.1
Introduction
Hemoglobin
genes are ancient, dating back perhaps as far as the origins of cellular
life. The familiar class of
hemoglobins used for oxygen transport illustrates only one function of
hemoglobins. This chapter will
review some of the principle events in the evolution of vertebrate globin gene
clusters within the context of their long history. This evolutionary framework provides some insights into
important issues such as the origin and function of the locus control regions,
the contrasting chromatin structure of alpha- and beta-like globin gene clusters,
and the prospects for targeting delta- or gamma-globin gene expression in
therapies for beta-globin gene defects.
5.2.
Broad distribution of hemoglobins in the biosphere
Hemoglobins
similar to human Hb A are found in erythrocytes of all vertebrates (Dickerson
and Geis 1983). Each is a
heterotetramer with two subunits related to the alpha-globin subfamily
(referred to here as alpha-like globins) and two subunits related to the
beta-globin subfamily (beta-like globins). The globin polypeptides bind heme, which in turn allows the
hemoglobin in erythrocytes to bind oxygen reversibly and transport it from the
lungs to respiring tissues. In all
species studied, different alpha-like and beta-like globins are synthesized at
progressive stages of development to produce hemoglobins characteristic of
primitive (embryonic) and definitive (fetal and adult) erythroid cells (Bunn
and Forget 1986). However, the
vertebrate hemoglobins comprise only a small part of the hemoglobin family
(Fig. 1). A close relative, the
monomeric myoglobin, stores oxygen in tissues such as muscle (Wittenberg and
Wittenberg 1987). As illustrated
by the summary phylogenetic tree in Fig. 1, the amino acid sequences of the
alpha- and beta-globins and myoglobin are related to each other, indicating a
common ancestor in early vertebrates approximately 500 million years ago
(Goodman et al. 1987). The
3-dimensional structure of myoglobin was one of the first solved, revealing a
series of alpha-helices that form the hemepsilon-binding pocket. This structure, the globin fold, is
seen in myoglobin, alpha-globin, and beta-globin; it is characteristic of all members of the hemoglobin family
of proteins (Dickerson and Geis 1983).
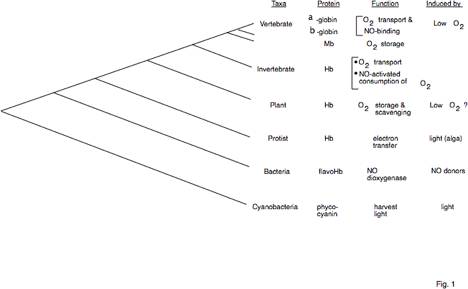
Fig. 1. Broad distribution and diverse
functions of hemoglobins. The
phylogenetic tree on the left is a summary of trees generated by aligning amino
acid sequences of hemoglobins from species representative of each taxa (using
CLUSTAL W) and computing trees based on parsimony (PAUP) and analysis of
distance measures by Neighbor joining and UPGMA. The latter two used the MEGA suite of programs (Kumar et al.
1993). Trees of the same topology
were generated by all three methods.
The summary tree shows that topology but is not drawn to scale. This and
subsequent trees indicate the relative time of the divergences; nodes more to the left indicate a
relatively earlier time. Functions
and induction agents are also listed.
More details and references have been reviewed (Hardison 1998).
Hemoglobins
are also used for oxygen transport in invertebrates (Riggs 1991; Dixon et al.
1992; Sherman et al. 1992). Many
non-vertebrates have gigantic extracellular hemoglobins, in some species formed
by as many as 200 monodomain subunits in a multimeric protein, and in others by
covalent linkage into very long polypeptide chains (reviewed in Terwilliger
1998). The invertebrate
hemoglobins are homologous to the vertebrate hemoglobins, and they form a
distinct branch in a phylogenetic tree of hemoglobins (Fig. 1). Hemoglobins are present in plants, both
the leghemoglobins with specialized functions in root nodules (Brisson and
Verma 1982; Appleby 1984) as well as the broadly distributed, nonsymbiotic
hemoglobins (Andersson et al. 1996).
Interestingly, the genes for plant and invertebrate hemoglobins have a
similar structure. Both groups of
genes have three introns separating four exons, with at least two introns in
identical positions (Fig. 2). The
similarities in gene structure and the amino acid sequences of the encoded
proteins strongly support the hypothesis of a common ancestor to both groups of
hemoglobins, showing that the evolutionary history of hemoglobin genes predates
the divergence of plants and animals, roughly 1.3 billion years ago (Feng et
al. 1997). It is likely the middle
intron was lost prior to the divergence of the vertebrate globin genes, all of
which have only two introns.
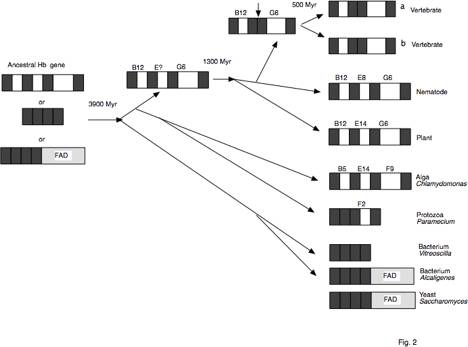
Fig. 2. Intron/exon structure during evolution
of hemoglobin genes.
The structures
of illustrative contemporary hemoglobin genes are shown on the right, with
exons denoted by dark boxes and introns by white boxes. The position in the hemoglobin
a-helical structure of the amino acid encoded at the site of interruption is
indicated over the intron, and the loss of the central intron in the ancestor
to vertebrates is marked by a vertical arrow. The evolutionary pathway is indicated by the other
arrows. This "tree" is a
gene tree, and grouping of of a yeast hemoglobin gene with bacterial hemoglobin
genes may reflect a horizontal gene transfer. Estimated times of divergence in millions of years (Myr) are
given at selected nodes.
Given
that the hemoglobins in the major groups of multicellular organisms - plants,
invertebrates and vertebrate animals - are used for storage and transport of oxygen,
one might have expected hemoglobins to be absent from unicellular
organisms. It was thought that
simple diffusion was sufficient to provide adequate oxygen inside the cells of
unicellular, freepsilon-living organisms. However, hemoglobins have now been characterized
in several species of eubacteria,
the fungus Saccharomyces cerevisiae, and protists
such as the alga Chlamydomonas and the protozoan Paramecium
(reviewed in Hardison 1996; Hardison 1998) These hemoglobins from unicellular organisms appear to play
roles distinctly different from those of vertebrate hemoglobins. The familiar functions in oxygen
transport and storage require reversible binding of oxygen, and that occurs only when the iron in
the heme stays in the reduced (+2) oxidation state, i.e. when it is a ferrous
ion. Biochemical analysis has
shown that hemoglobins from Chlamydomonas (Couture and
Guertin 1996), Saccharomyces (Zhu and Riggs 1992) and the bacterium Alcaligenes
(Cramm et al. 1994) can participate in electron transfer reactions in
vitro, with the
hemepsilon-bound iron changing cyclically between the +2 and +3 oxidation
states. The latter two hemoglobins
are actually two-domain proteins, one binding heme and the other binding flavin
cofactors, which usually plays a role in redox reactions. Also, the hemoglobin
from Vitreoscilla can
serve as a terminal electron acceptor during respiration in vivo (Dikshit et al. 1992).
Recent studies clearly show that the
hemoglobins in unicellular organisms have enzymatic functions and are not
oxygen-transporting proteins. The flavohemoglobins from the enteric bacteria Escherichia
coli and Salmonella
typhimurium (Crawford
and Goldberg, 1998; Gardner et al., 1998; Hausladen et al., 1998) and from yeast (Liu
et al., 2000) are
enzymes protecting these microorganisms from the highly reactive free radical
compound, nitric oxide. Each of these flavohemoglobins is a nitric oxide
dioxygenase, catalyzing the conversion of nitric oxide to nitrate. Other functions
have also been proposed for bacterial hemoglobins. For instance, the hemoglobin
from Vitreoscilla can
serve as a terminal electron acceptor during respiration in vivo (Dikshit et al., 1992).
Hemoglobins
involved in catalytic conversions of nitric oxide and oxygen are not limited to
microorganisms. A hemoglobin found in the perienteric fluid of the parasitic
worm Ascaris lumbricoides
also catalyzes reactions between oxygen and nitric oxide, producing nitrate (Minning
et al., 1999). However, the chemical mechanism is different from that of
the microbial flavohemoglobins, and Mining et al. (1999) propose that this
hemoglobin functions to remove oxygen from the perienteric fluid via a series
of reactions driven by nitric oxide.
In mammals, hemoglobins not only
transport oxygen, but they also help regulate nitric oxide levels. Gow et al. (1998) show that at physiological concentrations, nitric oxide
will bind to a cysteine in hemoglobin to form S-nitrosohemoglobin. This binding
is favored in oxyhemoglobin, and retains the bioactivity of nitric oxide.
Nitric oxide can subsequently be released from deoxyhemoglobin (Jia
et al., 1996; Stamler et al., 1997). Since nitric oxide is a major regulator of blood pressure,
these new findings indicate that hemoglobin is involved in the control of blood
pressure in ways that may facilitate efficient delivery of oxygen to tissues.
Furthermore, the interplay between binding of oxygen and nitric oxide to
hemoglobin and effects on vasodilation and constriction may have therapeutic
applications (e.g., Bonaventura et al., 1999; Gladwin
et al., 1999; Nagel, 1999).
The
variety of functions now found for hemoglobins raises the issue of whether the
microbial proteins are truly homologous to the hemoglobins from plants and
animals. The amino acid sequence
comparisons certainly support a common ancestor to all these sequences, as
illustrated in the summary phylogenetic tree (Fig. 1). Despite the low percent identity (e.g.
25%) between the more dissimilar members of the family, different types of
phylogenetic analysis generate trees of the same topology. The threepsilon-dimensional structures
strongly support the conclusion that all these hemoglobins share a common
ancestor. The structures of the
bacterial hemoglobins from Vitreoscilla (Tarricone et
al. 1997) and Alcaligenes
(Ermler et al. 1995) both have the globin fold first characterized in
vertebrate myoglobin. Indeed,
hemoglobins may be part of a larger family of hemoproteins. For instance, the light harvesting biliprotein, C-phycocyanin, from the
cyanobacterium Mastigocladus laminosus has a threepsilon-dimensional structure very similar to
that of a globin (Schirmer et al. 1985).
Although this is not a heme binding protein per se, it does bind a
linear tetrapyrrole pigment derived from heme. The structural comparisons
indicate that genes for at least some other hemoproteins share a common
ancestor with hemoglobin genes (Fig. 1).
These
observations all indicate that the gene encoding hemoglobin is truly ancient,
i.e. it appears to have been present in the ancestor to eubacteria and
eukaryotes, which is the earliest proposed divergence since the origin of
cellular organisms. This
divergence has been dated at approximately 3.9 billion years ago (Feng et al.
1997). At this early time, very
little oxygen was present in the earth’s atmosphere. Hence the primordial
function of hemoglobin may have had little to do with molecular oxygen (Hardison,
1998). The enzymatic functions of hemoglobins found in
contemporary microorganisms and nematodes, involving nitric oxide metabolism,
provide some insight into the early functions of hemoglobins (Durner
et al., 1999). Minning et al. (1999) describe a scenario in which hemoglobins present in
contemporary bacteria, which catalyze the enzymatic detoxification of nitric
oxide, represent an ancestral function. These ancestral hemoglobins may have
evolved into enzymes that catalyze the nitric oxidepsilon-mediated consumption
of oxygen, as now observed as a “deoxygenase” in Ascaris. This “deoxygenase” may have evolved
into contemporary mammalian hemoglobins, with their limited enzymatic function
but the ability to bind and transport both oxygen and nitric oxide.
Because
the separation between archaebacteria and eukaryotes appears to have occurred
after the divergence of eubacteria from eukaryotes (both of which have
hemoglobins), one may anticipate finding homologs to hemoglobins in archae as
well. An automated analysis of
genome sequences has included an archaebacterial gene (from Methanococcus
jannaschii) in an
orthologous group (Tatusov et al. 1997) containing proteins related to
hemoglobins (see http://www.ncbi.nlm.nih.gov/COG/). Further investigation of this and other archaebacterial
genes related to hemoglobins, revealed from the whole genome sequencing, should
provide even more insights into the origin and range of functions of hemoglobins.
An
issue that has received much attention is the age of the introns, and whether
they serve to separate genes into exons that encode distinct protein domains
(Gilbert 1978). The three introns
of globin genes in plants and invertebrates, dating back approximately 1.3
billion years, are between the segments of the gene encoding measurable domains
of protein structure (Go 1981).
This has lent support to the model that introns are old, and are the
remnants of a process that combined exons to generate genes with new structures
and functions (Gilbert 1978).
However, the hemoglobin genes from protists have introns in positions
unique to many of the species, and those of eubacteria have no introns (Fig.
2). Attempts to explain this
degree of heterogeneity as the result of differential loss of introns require a
very large number of introns to be proposed in the ancestral gene. An alternative explanation is that at
least some of the introns in the protist hemoglobin genes arose by insertion of
new introns in each lineage, consistent with the "introns late" model
(Stoltzfus et al. 1994). Thus it
seems unlikely that all the introns in contemporary hemoglobin genes were
present in the ancestral gene (i.e. preceding the divergence of eubacteria, archaebacteria
and eukaryotes), and hence the "introns early" hypothesis is not
adequate to explain all of the introns.
However, this does not rule out the possibility that some introns,
perhaps those still in hemoglobin genes in multicellular organisms, were in the
ancestral gene.
5.3.
Evolution of alpha- and beta-globin gene clusters in vertebrates
Human
hemoglobins are encoded at two separate loci, the beta-like globin gene cluster
on chromosome 11p15.5 (Deisseroth et al. 1978) and the alpha-like globin gene
cluster close to the terminus of chromosome 16p (Deisseroth et al. 1977). As shown in Fig. 3, the genes in each
cluster are in the same transcriptional orientation and are arranged in the
order of their expression during development, with the active beta-like globin
genes arranged 5'-epsilon (embryonic)-Ggamma (fetal)-Agamma (fetal)-delta
(minor adult)-beta (major adult)-3' (Fritsch et al. 1980) and the active
alpha-like globin genes arranged 5'-zeta (embryonic)-alpha2 (fetal and adult)-alpha1
(fetal and adult)-3' (Lauer et al. 1980).
[Lower levels of the gamma- and alpha-globins are also produced in
embryonic red cells.] This section
reviews some of the key events in the evolution of this arrangement of globin
genes.
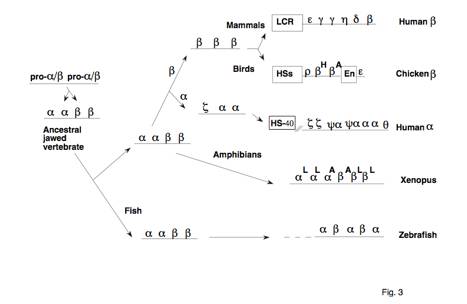
Fig. 3. Evolutionary pathways for alpha- and
beta-globin gene cluters in vertebrates.
Each gene is indicated simply by a Greek letter. Contemporary gene clusters are on the
right (Hardison 1991 and references therein), and the deduced course of
evolution to them is shown by a series of arrows. The ancestor to alpha- and beta-globin genes is indicated as
pro-alpha/beta. LCR = locus
control region, HS-40 = the distal major control region of mammalian
alpha-globin genes, HSs = DNase hypersensitive sites, En = enhancer.
The
effective transport of oxygen between tissues by hemoglobin is accomplished by
highly cooperative binding of oxygen when its concentration is high (e.g. in
the lungs), followed by cooperative dissociation when its concentration is low
(e.g. in respiring tissues in the periphery). In vertebrates, this cooperativity is accomplished by the
interactions between the alpha- and beta-globin subunits of hemoglobin (see
chapter 8 by M. Perutz).
Vertebrate hemoglobins are kept at high concentrations inside
erythrocytes, specialized cells devoted to the task of oxygen transport. Thus the divergence of the ancestral
globin gene into the alpha-globin and beta-globin genes (Fig. 3), and
expression of these genes at a high level only in erythroid cells, were key
steps in the evolution of cooperativity in hemoglobin and efficient oxygen
transport in vertebrates. These
goals have been accomplished by different mechanisms in other evolutionary
lineages. For instance, the basis
for cooperativity in non-vertebrate hemoglobins is quite different, in many
cases involving reversible dissociation of hemoglobin subunits upon oxygenation
(Riggs 1998).
Vertebrate
alpha- and beta-globin genes likely arose by the duplication and subsequent
divergence of an ancestral globin gene in early vertebrates. This would have generated a linked set
of alpha- and beta-globin genes (Fig. 3), which is the arrangement seen in
contemporary globin gene clusters of the teleost zebrafish Danio rerio
(Chan et al. 1997) and in the amphibian Xenopus (Hosbach et al. 1983). The alpha-globin gene cluster is thought
to have separated from the beta-globin gene cluster prior to the divergence of
birds and mammals, since these gene clusters are on separate chromosomes
in both groups of animals
(Deisseroth et al. 1976; Hughes et al. 1979). Gene duplication and divergence continued independently in
each of these lineages to generate the contemporary gene clusters. This is illustrated by the avian and
mammalian beta-globin gene clusters, which contain multiple genes expressed
differentially in development (Fig. 3).
In both species the epsilon- globin gene is expressed in embryos and the
beta-globin gene is expressed in adults.
However, the sequence of each chicken beta-like globin gene is equally
similar to each human gene (Goodman et al. 1987; Reitman et al. 1993), so that,
for example, chicken epsilon-globin is no more similar to human epsilon-globin
than to human beta-globin. This
indicates that the gene duplications generating these beta-globin gene clusters
occurred after the species diverged.
Much
is now known about the organization of alpha- and beta-globin gene clusters in
contemporary mammals, which can be understood in terms of descent from common
gene clusters in an ancestral eutherian mammal. Fig. 4 shows beta-globin gene clusters in species from five
orders of eutherian mammals and a marsupial. Analysis of DNA sequences showed that a given globin gene in
one species is usually more related to a gene in another mammal than to other
globin genes in the same species (Hardison 1983; Goodman et al. 1984; Hardies
et al. 1984; Hardison 1984; Townes et al. 1984). This indicated that these genes are orthologous, i.e., they are similar because of
descent from the same gene in the last common ancestor to the two species. The exceptions to this observation could
be explained by gene duplications within a single mammalian lineage, e.g. the
duplication of gamma-globin genes in the ancestor to simian primates to produce
paralogous genes
(similar because of duplication of the ancestral gene), such as the Ggamma- and
Agamma-globin genes in humans (Shen et al. 1981; Fitch et al. 1991). Finding orthologs to epsilon-, gamma-,
eta-, delta- and beta-globin genes, in that order, in virtually all eutherian
mammals suggested that the ancestral eutherian had at least this set of genes
(Fig. 4). This hypothesis was
strongly supported by the observation of substantial regions of sequence
similarity outside the coding regions of the genes, in the introns and flanking
regions (Hardies et al. 1984; Hardison 1984; Margot et al. 1989; Shehee et al.
1989; Hardison and Miller 1993).
As will be discussed more extensively below, some but not all of these
matching sequences are strong candidates for regulatory function. The long regions of matching sequences
outside functional regions, and thus not subject to any obvious selection, were
key observations in establishing this model for evolution of the mammalian
globin gene clusters. Deletions,
conversions and duplications of both single genes and blocks of genes have
occurred in each mammalian order to generate the current gene clusters
(reviewed in Collins and Weissman 1984; Hardison 1991).
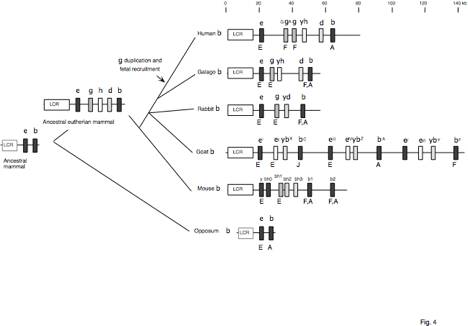
Fig. 4. Pathways to contemporary mammalian
beta-globin gene clusters. Genes
are indicated by boxes, and orthologous genes have the same type of fill. The presumptive presence of an LCR in
marsupials is indicated by the grey outline (R. Hope, personal
communication). The stage of
expression is indicated as E = embryonic, F = fetal, and A = adult. References are in the text and in
reviews (Collins and Weissman 1984; Hardison 1991). a=alpha, b=beta, z=zeta,
e=epsilon, g=gamma, d=delta, r=rho, q=theta
The
proposed epsilon-gamma-eta-delta-beta globin gene cluster in the ancestral
eutherian mammal was generated by earlier gene duplications. Estimates based on rates of divergence
indicated that the epsilon-, gamma- and eta-globin genes arose from
duplications of one ancestral gene, whereas the delta- and beta-globin genes
arose by duplication of a different gene, perhaps prior to the divergence of eutherian
and metatherian (marsupials and monotreme) mammals (Goodman et al. 1984;
Hardison 1984). This prediction
was verified by genomic analysis of marsupials (Koop and Goodman 1988; Cooper
et al. 1996), which have two genes in their beta-globin gene clusters, one most
related to eutherian epsilon-globin genes and the other most related to
beta-globin genes. Thus the model
shown in Fig. 4 is robust, in that it has been supported not only by deductions
from analysis of contemporary species, but also by tests of predictions made by
the model.
One
important ramification of this model is that orthologous genes have not
retained the same time of expression during development in all mammalian
orders. The gamma-globin gene in
most mammals is expressed in embryonic erythroid cells, but in simian primates,
including humans, it is expressed predominantly in fetal erythroid cells. Concomitantly with the fetal
recruitment of the gamma-globin gene, expression of the beta-globin gene has
been delayed in higher primates so that in humans it is expressed primarily in
post-natal life. In other mammals,
the beta-globin gene is expressed in both fetal and adult erythroid cells. In
goats, the gamma-globin gene has been deleted, and subsequent expansion of the
gene cluster by triplication of a four-gene set (Townes et al. 1984) allowed expression of the resulting
paralogous beta-globin genes in fetal life (betaF), adult life (betaA)
or under conditions of erythropoietic stress (betaC). The delta-globin
gene is expressed at low levels in adult humans, but is silent in some mammals,
and is expressed at high levels in others. In contrast, the epsilon-globin gene in each mammalian
species is expressed only in embryonic erythroid cells derived from the yolk
sac. Within the beta-globin gene
clusters of mammals, conservation of stage-specific expression is seen only for
this gene, which is located closest to the distal locus control region (LCR,
see below and chapter 6 by B. Forget).
Perhaps the embryonic restriction of epsilon-globin gene expression is
related to this spatial relationship, with active expression in the embryonic
lineage due to its proximity to the LCR, followed by silencing in the fetal and
adult (definitive) lineage of erythroid cells (see chapter 14 by G. Stamatoyannopoulos). Both the proximity to the LCR and the
embryonic restriction to expression is conserved in all mammalian
epsilon-globin genes examined.
The
beta-globin gene clusters of humans and mice are embedded within a large
cluster of olfactory receptor genes, or ORGs (Bulger
et al., 1999). This arrangement suggests that the beta-globin genes
transposed into a prepsilon-existing array of ORGs. A related ORG is found on
the 3’ side of the chicken beta-globin gene cluster (Bulger
et al., 1999), but an erythroid-specific folate receptor gene is located
on the 5’ side, separated from the beta-globin gene cluster by an insulator (Prioleau
et al., 1999). The 3’ breakpoints of at least two deletions causing
hereditary persistence of fetal hemoglobin (HPFH) in humans are close to ORGs
located 3’ to the beta-globin genes (Kosteas
et al., 1997; Feingold et al., 1999). The ORG close to the HPFH-1 breakpoint is in an open
chromatin domain in human erythroid cells (Elder
et al., 1990). Enhancer sequences from this ORG are brought in proximity
to the gamma-globin genes by the HPFH-1 deletion, and this may play a role in
the increased expression of gamma-globin genes in adults carrying this deletion
(Feingold and Forget, 1989).
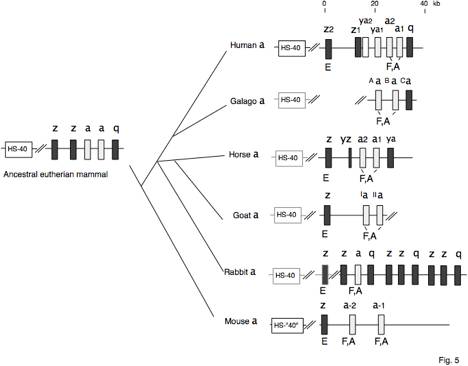
Fig. 5. Pathways to contemporary mammalian
alpha-globin gene clusters. Genes
are indicated by boxes, and orthologous genes have the same type of fill. The presumptive presence of a homolog
to HS-40 (the distal major control region) in mammals besides human and mouse
is indicated by the grey outline.
References are in the text and in reviews (Collins and Weissman 1984;
Hardison 1991; Hardison and Miller 1993). a=alpha, b=beta, z=zeta, e=epsilon,
g=gamma, d=delta, r=rho, q=theta
The
evolution of alpha-globin gene clusters in contemporary mammals is not as well
understood, in part because less information is available on gene organization
and sequence in non-human mammals, and in part because the rate of sequence
change in this gene cluster appears to be higher than in the beta-globin gene
cluster (Hardison et al. 1991).
Fig. 5 summarizes the arrangement of alpha-like globin gene clusters in
representatives of 5 orders of eutherian mammals. Orthologous relationships have been assigned primarily on
the basis of DNA sequence matches outside the genes (Hardison and Gelinas 1986;
Sawada and Schmid 1986; Wernke and Lingrel 1986; Flint et al. 1988), even
though such matches are considerably more limited than in the mammalian
beta-globin gene clusters. Since a
variant of the arrangement 5'-zeta-zeta-alpha-alpha-theta-3' is found in all
contemporary mammals examined, it is likely that these genes were present in
this order in the gene cluster of the ancestral eutherian mammal. The timing of expression is
well-conserved among these mammals.
The zeta-globin genes are expressed only in embryonic erythroid cells,
whereas the alpha-globin genes are expressed in all erythroid cells, albeit at
lower levels at the embryonic stage (Rohrbaugh and Hardison 1983; Leder et al.
1985; Peschle et al. 1985). The
theta-globin genes are still not well understood. The human theta-globin gene is transcribed at low levels but
does not encode any known polypeptide found in human hemoglobins (Hsu et al.
1988; Kim et al. 1989; Leung et al. 1989). It is a feature of every mammalian alpha-like globin gene
cluster examined (Fig. 5), and given that gene deletions can occur in this
locus (see below), one would anticipate the loss of nonfunctional genes in at
least some mammalian lineages. The
retention of the theta-globin genes is suggestive of some functional
importance, but perhaps not for encoding a globin polypeptide.
Although
no examples of recruitment for expression at different developmental stages are
seen in the alpha-like globin gene clusters, some genes have lost their
function during evolution. In
particular, based on upstream sequence matches, the human ya1-globin pseudogene
appears to be orthologous to an active alpha-globin gene in goats and horse
(Fig. 5). The inactivation of ya1-gene
is accompanied by the loss of a CpG island that encompasses its homologs (Bird
et al. 1987). The orthologous
relationships in Fig. 5 indicate that the a2- and a1-globin genes in humans
result from a duplication only in primates, i.e. separate from the duplication
proposed to generate the pair of alpha-globin genes in the ancestral eutherian
mammal. The more recent
duplication in primates has left a long region of sequence similarity
surrounding the alpha-globin genes (Hess et al. 1984), and unequal cross-overs
within that region of homology is the cause of some forms of
alpha-thalassemia. Not all mammals
have retained a pair of active alpha-globin genes. Rabbits are the exception, with only one alpha-globin gene
(Cheng et al. 1986). Curiously,
this gene cluster has expanded by block duplications of a zeta-zeta-theta gene
triad (Cheng et al. 1987), similar to the expansion of the beta-like globin
gene cluster in goats (Townes et al. 1984) (Fig. 4).
All
the vertebrate globin gene clusters examined to date encode subunits of
hemoglobins differentially expressed in embryonic and adult erythroid cells
(Fig. 3). Likewise, hemoglobin
synthesis is developmentally regulated in some invertebrates (Terwilliger 1998)
and different plant leghemoglobins are made at progressive stages of nodulation
(Hyldigamma-Nielsen et al. 1982; Lee et al. 1983). Even species as distant from human as Chlamydomonas
(Couture et al. 1994) and Paramecium (Yamauchi et al. 1995) have multiple
hemoglobin genes. Thus the ability
to express different hemoglobins at particular developmental stages, i.e.
hemoglobin switching, is very old, predating the plant-animal divergence, and
possibly being much older. In Fig.
3, multiple globin genes are shown in the ancestral gene clusters. It is likely that they were
differentially expressed during development in these ancestral species.
5.4 Differences in genomic context and
regulation of the mammalian alpha-globin and beta-globin gene clusters
The
separation of alpha-globin and beta-globin gene clusters to different
chromosomes has allowed them to diverge into strikingly different genomic
contexts, with paradoxical consequences for our understanding of their regulation. Given that all contemporary vertebrates
have developmentally regulated hemoglobin genes encoding proteins used for
oxygen transport in erythrocytes, it would have been reasonable to expect that
the molecular mechanisms of globin gene regulation would be conserved in
vertebrates. Certainly, the
coordinated and balanced expression of alpha- and beta-globin genes to produce
the heterotypic tetramer alpha2beta2 in erythrocytes should be a particularly
easy aspect of regulation to explain.
Since the two genes would have been identical after the initial
duplication in the ancestral vertebrate, with identical regulatory elements, it
is parsimonious to expect
selection to keep the regulatory elements very similar.
However,
much has changed between the alpha-like and beta-like globin gene clusters
since their duplication. Not only
are they now on separate chromosomes in birds and mammals, but in mammals they are in radically
different genomic contexts (Fig. 6).
The beta-globin gene clusters are A+T rich, with no CpG islands
(reviewed in Collins and Weissman 1984), whereas the alpha-like globin gene
clusters are highly G+C rich, with multiple CpG islands (Fischel-Ghodsian et
al. 1987). Tissue-specific gene expression
is frequently correlated with an increased accessibility of the chromatin only
in expressing cells, and hence "opening" of a chromatin domain is a
key step in activation of many tissue-specific genes. This is the case for beta-like globin genes of mammals
(Groudine et al. 1983; Forrester et al. 1990), but not the alpha-like globin
genes, which are in constitutively open chromatin (Craddock et al. 1995). Thus the mammalian alpha-globin genes
have several characteristics associated with constitutively expressed,
"housekeeping" genes.
Additionally, the alpha-globin genes are replicated early in S phase in
all cells (a time when most expressed genes are replicated), whereas
beta-globin genes are replicated early in S phase only in cells expressing them
(Calza et al. 1984; Dhar et al. 1988).
In keeping with the presence of CpG islands, the alpha-globin gene
cluster is not methylated in any cell types (Bird et al. 1987), whereas the
beta-globin gene cluster is subject to tissue-specific DNA methylation (van der
Ploeg and Flavell 1980). Thus the
strikingly different genomic contexts of the two gene clusters affect several
aspects of DNA and chromatin metabolism, including timing of replication,
extent of methylation, and the type of chromatin into which the loci are
packaged. Rather than selecting
for similarities to insure coordinate and balanced expression, the processes of
evolution at these two loci have made them quite different.
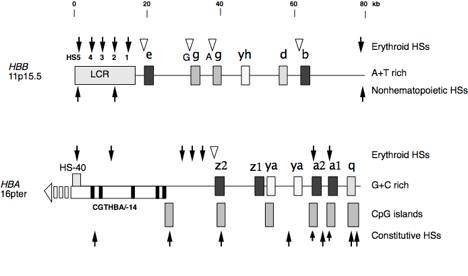
Fig. 6. Differences in chromatin structure
between alpha- and beta-globin gene clusters of humans. Globin genes and distal
control regions are shown as filled boxes. HS-40 is located within an intron (white box) of the -14
gene (exons of this gene are shown as black boxes), located upstream of the
zeta-globin gene and transcribed in the opposite orientation. Developmentally stable DNase
hypersensitive sites (HSs) are shown as filled arrows, and those that occur at
specific developmental stages (when the associated gene is expressed) are shown
as white triangles. CpG islands
are shown as boxes with horizontal lines.
None are in the beta-globin gene.
HBB=
beta-globin gene, HBA
= alpha-globin gene. References
are in the text. a=alpha, b=beta, z=zeta, e=epsilon, g=gamma, d=delta, r=rho,
q=theta
The
alpha and beta-globin gene clusters also have important differences in their cis-regulatory elements. Both have distal control elements,
called the locus control region (or LCR) for beta-globin genes (reviewed in
Grosveld et al. 1993; Hardison et al. 1997b) and HS-40 for the alpha-globin genes
(Higgs et al. 1990), that are required for higeta-level expression of the
respective globin genes in transgenic mice, independently of the position of
chromosomal integration. However,
they differ in the range of functions associated with them. The beta-globin LCR
is required for tissue-specific chromosomal domain opening (Forrester et al.
1987; Forrester et al. 1990), whereas no such function has been implicated for
HS-40, as expected for a regulator in constitutively open chromatin. The distal regulatory elements also
differ dramatically in size, with the beta-globin LCR containing 17 kb with 5
DNase hypersensitive sites in chromatin (Tuan et al. 1985; Forrester et al.
1987; Grosveld et al. 1987; Dhar et al. 1990), compared to about 0.4 kb and a
single DNase hypersensitive sites in chromatin for the alpha-globin HS-40
(Jarman et al. 1991).
Indeed,
the alpha-globin HS-40 is most similar to a single hypersensitive site, HS2,
from the beta-globin LCR. Both
will confer inducible, high level expression on reporter genes in transfected
cells (e.g. Tuan et al. 1985; Ney et al. 1990; Pondel et al. 1992; Ren et al.
1993), in addition to their effects in transgenic mice (Fraser et al. 1990;
Higgs et al. 1990; Morley et al. 1992).
As illustrated in Fig. 7, these two enhancers share binding sites for
some, but not all, transcription factors (e.g. Talbot et al. 1990; Jarman et
al. 1991; Strauss et al. 1992; Reddy et al. 1994). Both contain maf-response elements, or
MAREs (Motohashi et al. 1997), to which transcriptional activator proteins of
the basic leucine zipper class can bind.
A particular subfamily of proteins related to AP1, such as NFE2,
LCRF1/Nrf1, and Bach1, bind to this site (reviewed in Orkin 1995; Baron 1997). All are heterodimers containing a Maf
protein as one subunit, which is the basis for the name. Other binding sites in common are GATA,
to which GATA1 and its relatives bind (Evans et al. 1990), and the CACC motif,
to which a family of Zn-finger proteins including EKLF can bind (Miller and
Bieker 1993) . These binding sites
are occupied in vivo
(Strauss et al. 1992; Reddy et al. 1994), and all have been shown to contribute
to the function of the enhancers (Strauss et al. 1992; Caterina et al. 1994;
Reddy et al. 1994; Rombel et al. 1995).
Other functional sites, such as the E boxes in HS2 (Lam and Bresnick
1996; Elnitski et al. 1997), are not found in common. Each of these binding sites is conserved in homologous
regulatory elements in mammals (reviewed in Gourdon et al. 1995; Hardison et
al. 1997b). In general, binding
sites for many of the proteins implicated in activation of globin gene
expression are present in both HS-40 and HS2 of the beta-globin LCR, but their
number and arrangement differs in the two enhancers (Fig. 7). It is currently not possible to assess whether
this limited similarity occurs via divergence from a common ancestral
regulatory element or by convergence from different ancestral elements.
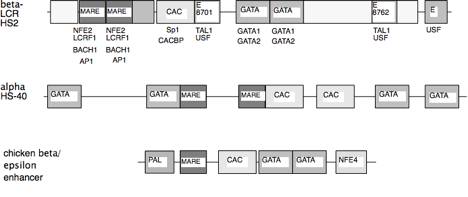
Fig. 7. Conserved motifs in distal regulatory
elements. Similar protein binding
sites have the same fill, and proteins implicated in acting at a given site are
listed below that motif in the beta-LCR HS2 line; the same proteins have been
implicated at similar motifs in the other regulatory elements. Boxes without labels are conserved
sequences of untested function.
The
proximal regulatory elements also differ in important ways between alpha-globin
and beta-globin genes (Fig. 8).
The promoters do contain two binding sites in common - the TATA motif,
to which the general transcription factor TFIID binds, and the CCAAT motif, to
which several families of trans-activators,
such as CP1, can bind
(Efstratiadis et al. 1980).
However, the other protein binding sites are completely different
between the human alpha and beta-globin genes (deBoer et al. 1988; Rombel et
al. 1995). In addition, the CpG
island encompassing the 5' flanking region and much of the gene is a key
component of the cis-regulatory
elements for the alpha-globin gene of rabbits and humans, possibly through its
effects on chromatin structure (Pondel et al. 1995; Shewchuk and Hardison
1997); again, no CpG island is
found at any of the beta-like globin genes.
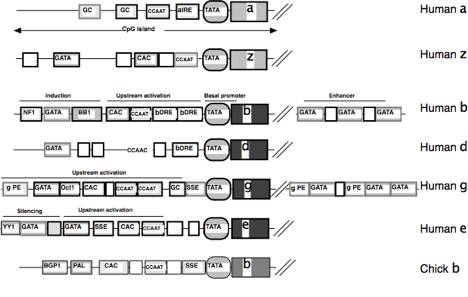
Fig. 8. Conserved features of globin gene
promoters. Binding sites for the
human globin genes are shown;
similar protein binding sites have the same fill. Those sites conserved in other mammals
have a dark outline, those not conserved have a grey outline. The figure is not drawn to scale, but relative
positions are indicated; the genes
themselves are truncated. The
chicken beta-globin gene promoter is shown for comparison; no information about evolutionary
conservation is presented for this gene.
This figure summarizes work from a large number of papers (e.g.
Efstratiadis et al. 1980; Lacy and Maniatis 1980; Hardison 1983; Antoniou et
al. 1988; Barnhart et al. 1988; deBoer et al. 1988; Wall et al. 1988; Martin et
al. 1989; Puruker et al. 1990; Stuve and Myers 1990; Macleod and Plumb 1991;
McDonagh et al. 1991; Yu et al. 1991; Gong and Dean 1993; Motamed et al. 1993;
Peters et al. 1993; Yost et al. 1993; Lloyd et al. 1994; Rombel et al. 1995). a=alpha,
b=beta, z=zeta, e=epsilon, g=gamma, d=delta, r=rho, q=theta.
The
differences in genomic context between alpha-globin and beta-globin genes have
been seen for non-human mammals as well (Bernardi et al. 1985), indicating an
origin prior to the divergence of eutherian mammals. The high G+C content and presence of CpG islands is
characteristic of alpha-globin gene clusters from goats (Wernke and Lingrel
1986), horse (Flint et al. 1988) and rabbit (Hardison et al. 1991), whereas the
beta-globin gene clusters from non-human mammals are rich in A+T (e.g. Margot
et al. 1989; Shehee et al. 1989).
The only apparent exception is the mouse alpha-globin gene cluster,
which to date has not been completely characterized. However, the sequenced mouse alpha-globin gene is not in a
CpG island (Nishioka and Leder 1979).
Indeed, the mouse genome shows a general depletion in CpG islands
(Antequera 1993; Matsuo et al. 1993).
The loss of the CpG island is correlated with a chromosomal
rearrangement that moved the alpha-globin locus from the terminus of a
chromosome, where it is in human (Flint et al. 1997) and rabbit (Xu and
Hardison 1991), to an internal location in mouse (Tan and Whitney 1993). It is possible that position of the
alpha-globin locus close to the end of the chromosome, in a large region high
in G+C content (Flint et al. 1997), is critical to maintenance of the CpG
islands.
Despite
these many differences between alpha-globin and beta-globin gene clusters in
mammals, the appropriate genes are still expressed coordinately between the two
loci, resulting in balanced production of alphalike and beta-like globins
needed for the synthesis of normal hemoglobins. The full mechanism that accomplishes this task still eludes
our understanding.
5.5.
Origin and location of the LCR in birds and mammals
In
contrast to the differences between the alpha and beta-globin gene clusters of
mammals, tissue-specific opening of a chromatin domain occurs in beta-globin
gene clusters of both mammals and birds.
In fact, the association between accessible chromatin and gene
activation was first made for the chicken globin genes (Weintraub and Groudine
1976). Further studies of the
avian gene clusters carefully mapped the limits of the open domain in chromatin
to a region of 33 kb, extending about 10 kb on each side of the set of 4 globin
genes (Clark et al. 1993; Hebbes et al. 1994). The limits of the open domain for the human beta-globin gene
cluster have not been determined.
The open domain at the 5' end encompasses at least the LCR, which
extends 20 kb 5' to the epsilon-globin gene, and an erythroid HS has been
mapped as far as 70 kb 3' to the beta-globin gene (Elder et al. 1990),
indicating an open domain of at least 150 kb.
LCRs
have been mapped in beta-globin gene clusters both in chickens and in all
mammals examined to date. The LCRs
in mammals are homologous (Moon and Ley 1990; Li et al. 1991; Hug et al. 1992;
Jimenez et al. 1992; Hardison et al. 1993; Hardison et al. 1997b; Slightom et
al. 1997), with long segments of high sequence similarity found both inside and
outside the cores of the LCR HSs (Fig. 9). All are located 5' to the epsilon-globin gene (Fig.
4). Sequences similar to HS2 and
HS3 of the beta-globin LCR are also found in marsupials and monotremes (R. Hope,
personal communication), indicating that the LCR is predates the divergence of
placental and nonplacental mammals, about 173 Myr ago (Kumar and Hedges
1998). In contrast, the major LCR
activity in chickens maps to an enhancer located between the betaA and
epsilon-globin genes (Reitman et al. 1990). Four additional HSs are 5' to the rho-globin gene, and
together they produce a modest enhancement in expression (Reitman et al. 1995). Despite their analogous location at the
5' ends of the gene cluster, they do not have the pronounced effects associated
with the HSs in the mammalian LCR.
In fact, comparison of the DNA sequences of the beta-globin gene
clusters between chicken and human fails to reveal any statistically
significant alignments outside the coding regions of some of the exons (Reitman
et al. 1993; Hardison 1998). Thus
no clear homologies are present in either the distal regulatory elements (LCRs
and enhancers) or in the promoters.
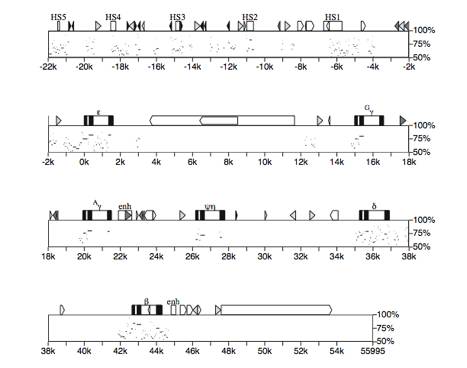
Fig. 9. Positions and degree of similarity of
alignments between human and mouse beta-globin gene clusters. This percent identity plot, or pip
(Hardison et al. 1997a), shows a detailed map of the human beta-globin gene
cluster along the top horizontal axis, with the positions of segments that
align with the mouse sequence shown as horizontal lines in the plot. The vertical position in the plot is
the percent identity of each aligning segment. The HSs in the LCR are appropirately labeled white boxes,
globin genes are shown with black exons and white introns, and the 3' enhancers
are labeled enh. Interspersed
repeats were identified using Repeat Masker (U. of Washington) and are
indicated as follows:white arrowed boxes = L1 repeats, black arrowed boxes = L2
repeats, light grey triangles = Alu repeats, black triangles = MIR repeats,
darker grey triangles or arrowed boxes = other repeats, like LTR repeats, and
DNA transposons. One factor in the
limited amount of matching sequences in the region between the delta- and beta-globin
genes is the insertion of an HMG14 pseudogene for the gene between betah3 and
the beta-major globin gene in mouse (S. Philipsen, personal communication).
Again
we are left with a conundrum. The
separation of birds and mammals is estimated to have occurred about 310 Myr ago
(Kumar and Hedges 1998), and thus the avian and mammalian beta-globin gene
clusters shared a common ancestor at that time. Concomitant with the independent duplications and divergence
of genes in each lineage to make the contemporary gene clusters (Fig. 3), any
remnant of common regulatory elements has been lost, at least at the level of
primary DNA sequence analysis.
Despite this, the major features of regulation, such as chromatin
domain-opening directed by a distal regulatory element and stage-specific
activation of globin gene promoters, have been retained in each lineage. Considering that these features were
likely present in the ancestor to birds and mammals, it is surprising that the
regulatory elements have diverged so much. However, enhancer sequences are conserved between mammals
and chickens at other loci, such as SCL/TAL1 (Gottgens
et al., 2000). Enhancers of Hox gene clusters are conserved between human and pufferfish
(Aparicio et al., 1995). A full explanation of the lack of homology in the regulatory
regions of the chicken and human beta-globin gene clusters awaits a better
understanding of their evolutionary history, which is still under study.
Even
though the pairwise alignments did not reveal sufficiently long matching
segments to be significant, a comparison of the protein-binding sites in known
regulatory regions shows that some of the same proteins are used in both
species. For instance, the chicken
b/e enhancer has a MARE, a CACC motif and a GATA site (Evans et al. 1990),
motifs also found in HS2 of the beta-globin LCR and the alpha-globin HS-40
(Fig. 7). All three have a group
of binding sites in the order MARE - CACC - GATA, but the number and spacing of
the sites differ in the three enhancers.
The inability to detect these similar regulatory regions using
nucleotide identities as the basis for a similarity score illustrates the need
to develop software that finds similar patterns of protein binding sites in
comparisons of DNA sequences.
5.6.
Conserved sequences in the proximal regulatory regions of mammalian alpha-like
and beta-like globin genes
Unlike
the absence of extragenic sequence matches in comparisons between beta-globin
gene clusters of chickens and humans, or between alpha-globin and beta-globin
gene clusters of humans, comparisons of homologous gene clusters between
eutherian mammals reveal many informative sequence matches. As illustrated in
Fig. 9 for a comparison of the DNA sequences of the human (Collins and Weissman
1984; Li et al. 1985) and mouse (Shehee et al. 1989) beta-globin gene clusters,
extensive matches are seen outside the coding regions, including the 5'
flanking regions and in the distal locus control region. Pairwise comparisons of the human gene
cluster with that from galago (Tagle et al. 1992) or rabbit (Margot et al.
1989) show even more sequence matches (Hardison and Miller 1993).. When sequences from several species are
aligned simultaneously, one can detect conserved blocks of sequences (Miller et
al. 1994), or phylogenetic footprints (Tagle et al. 1988; Gumucio et al. 1996),
that are clearly changing more slowly than the surrounding sequences. The well-conserved sequences are
reliable guides to functional regions (e.g. Gumucio et al. 1992; Gumucio et al.
1993; Elnitski et al. 1997).
The
multiple sequence alignments can be used to find conserved protein binding
sites throughout the mammalian beta- and alpha-globin gene clusters. A detailed map of the human beta-globin
gene clusters is shown in Fig. 10, along with the positions of all matches to
consensus binding sites for selected proteins. Those binding sites that are conserved in all available
sequences are denoted by longer vertical lines. This analysis illustrates dramatically the restriction of
conserved MAREs to the HSs 2 and 3 in the LCR. Conserved CACC motifs are also found in HS2 and HS3, and
conserved GATA sites are in four HSs of the LCR, with HS3 containing 3 such
sites. In keeping with its high
content of A+T, the beta-globin gene cluster has an abundance of GATA sites but
almost no GC boxes, which are binding sites for Sp1. Only a small subset of the matches to binding sites are
conserved in other mammals, and these conserved sites tend to be in the 5'
flanking regions (but as much as a 1000 bp away from the genes) and in the
LCR.
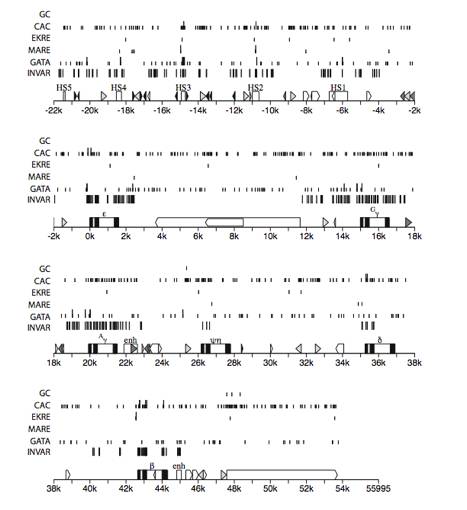
Fig. 10.
Distribution and conservation of sequence motifs throughout mammalian beta-globin
gene clusters. A detailed map of
the gene cluster is shown on the numbered line, using the same conventions as
in Fig. 9. Results of analysis of
a multiple alignment of beta-globin gene cluster sequences (available at the
Globin Gene Server, http://globin.cse.psu.edu) are shown above this map. The positions of invariant blocks at
least 6 columns long are on the line labeled INV (this is one criterion for
conserved sequences). Matches to
the following sequence motifs or their reverse complement are indicated as
labeled: GATA = WGATAR, MARE
(maf-response element) = TGASTCA, EKRE (EKLF-reponse element) = CCNCACCC, CACC
(core of site for binding Kruppel class Zn finger proteins) = CACC, GC (binding
site for Sp1) = CCGCCC. Those
motifs that are conserved in other species in the alignment (at least three of
them) are denoted by long
vertical lines; those that are not conserved are denoted by short vertical lines.
The
results of this type of analysis in the proximal regulatory regions of
mammalian alphalike and beta-like globin genes are summarized in Fig. 8. The conserved sites are outlined with
thicker lines. Only two binding
sites, the TATA box and CCAAT box (Efstratiadis et al. 1980) are found in all these globin
genes. A set of binding sites is
distinctive to each type of gene.
For instance, bDRF (Stuve and Myers 1990), EKLF (Miller and Bieker 1993;
Donze et al. 1995; Nuez et al. 1995; Perkins et al. 1995) and BB1-binding
protein (Antoniou et al. 1988; Macleod and Plumb 1991) have been implicated
only in the regulation of the beta-globin gene. All are conserved in mammals. EKLF binds to a CACC motif, and similar but distinctive CACC
motifs is also found in a comparable position in the 5' flanks of the gamma-
and epsilon-globin genes, again conserved in orthologous mammalian genes. The specificity of EKLF in regulating
the beta-globin gene raises the intriguing possibility that other, related
proteins binding to CACC motifs, perhaps active only at one developmental
stage, are regulating gamma- and epsilon-globin genes. A GATA site is conserved at about the
same position in both gamma- and epsilon-globin gene 5' flanking regions; these have been implicated in positive
regulation of the respective genes (Martin et al. 1989; Gong and Dean 1993).
Interestingly, the comparable region for beta-globin does not have a conserved GATA site (Hardison et al. 1994). Even conserved DNA sequence motifs in
paralogous genes may serve as binding sites for different proteins. A CCAAT motif located at about -80 in
all the vertebrate globin gene promoters can be bound by a heteromeric complex
called CP1, NF-Y or CBF (Hooft van Huijsduijnen et al. 1990 and references therein)
. However, preparations of CP1
bind much more strongly to the CCAAT box in the alpha-globin gene promoter than
in the beta-globin gene promoter (Cohen et al. 1986). Also, multiple additional proteins bind to the CCAAT box,
some of which have been implicated in the activation of beta-globin gene
expression (deBoer et al. 1988; Delvoye et al. 1993).
Thus
sequence alignments of these groups of orthologous genes in different eutherian
mammals reveal protein binding sites important for regulated expression. The
differences in the arrays of proteins functioning at epsilon-, gamma-, beta-
and alpha-globin genes indicate that a distinct battery of proteins functions
in the promoter for each type of gene.
Indeed, this is consistent with the observation that cis-acting sequences needed for
stage-specific regulation of expression map close to the genes (Trudel and
Costantini 1987 and references therein).
In contrast to the conservation of sites in the 5' proximal regions, the
enhancers located 3' to the Agamma-
and beta-globin genes (Bodine and Ley 1987; Wall et al. 1988; Puruker et al.
1990) are not well-conserved outside humans (Fig. 8), indicative of a function
peculiar to higher primates. One such function discussed in more detail below
is the expression of the gamma-globin genes in fetal life.
5.7.
Rise and fall of the delta-globin gene: new prospects for therapy
HbA2 (alpha2delta2) can inhibit the polymerization of
deoxy-Hb S in patients with sickle cell disease (Nagel et al. 1979; Poillon et
al. 1993). Since low levels of the
minor Hb A2 are present in all adult
erythrocytes (Heller and Yakulis 1969), strategies to increase synthesis of
delta-globin should have a pancellular effect on the decrease in sickling, a
possible advantage over the strategies to increase synthesis of gamma-globin,
which is expressed only in a subpopulation of erythrocytes. Examination of delta-globin gene
sequences in other mammals provides some important insights into sequence
changes that can activate or silence its expression.
The
evolutionary history of the delta-globin gene is complex, with recombinations
and mutations resulting in the loss, silencing or expression at a range of
levels in different mammals. Even
identifying beta-like globin genes that are orthologous to the human
delta-globin gene is complicated, because the human gene itself is a hybrid of
a 5' region closely related to the beta-globin gene fused to a 3' region that
is distinctive for the delta-globin gene (Spritz et al. 1980; Martin et al.
1983). This is the result of a
gene conversion event (Fig. 11), with the beta-globin gene serving as the donor
of the DNA sequences to the 5' region during the recombination (Martin et al.
1983). Thus for comparisons with other mammalian globin gene clusters, matching
sequences in the 3' portion are most useful for assignments of orthologous
relationships with the human delta-globin gene (Fig. 4). Further analysis reveals a striking
tendency for the delta-globin genes to undergo gene conversions with
beta-globin genes (Fig. 11); this
has occurred independently in the several primates, rabbits (Lacy and Maniatis
1980; Hardison and Margot 1984), and mice (Hardies et al. 1984).
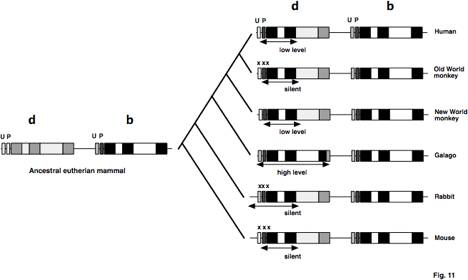
Fig. 11. Gene conversion, silencing and
activation of delta-globin genes during mammalian evolution. The ancestral pair of beta- and delta-globin
genes are shown with fills distinctive for the exons and introns of each
gene. The basal promoter is
indicated by the box labeled P, the uptream activating sequence indicated by
the box labeled U (shown in more detail in Fig. 8). The extent of each gene conversion is shown by the
doublepsilon-headed arrow. The x's
indicate mutations that silence the gene. d=delta, b=beta
Expression
of the delta-globin genes varies over a wide range in primates. In contrast to the low level expression
in humans, the delta-globin gene is silent in Old World Monkeys (Martin et al.
1983). It is expressed at a low
level in a New World Monkey, the spider monkey (Spritz and Giebel 1988), and at
a high level (up to 40% of total fetal and postnatal hemoglobin) in the
prosimian primates galago and tarsier (Tagle et al. 1991). In the galago, the delta-globin gene
has undergone an extensive gene conversion that effectively replaces it with
beta-globin gene sequences, i.e. extending from 800 bp 5' to the CCAAT box to
near the end of exon 3. This shows
that the delta-globin gene locus can be activated to high level expression, if
the promoter is appropriately modified (in this case replacing the delta-globin
gene promoter with the beta-globin gene promoter).
Comparison
of the promoter sequences between the silenced or low-level expression
delta-globin genes and the highly expressed beta-globin genes have revealed
several mutations associated with low level expression (Lacy and Maniatis 1980;
Martin et al. 1983; Tang et al. 1997).
Two prominent changes in the delta-globin gene promoters are the
mutation of the CCAAT box to a CCAAC and the loss of the proximal CACC box,
which is the binding site for EKLF in beta-globin gene promoters (Fig. 8). Site-directed mutagenesis experiments
have shown that these alterations in the delta-globin gene promoter do cause
lower level expression. Directed
mutations that restore the CCAAT box or insert a CACC motif will activate
delta-globin reporter gene expression in transfected erythroid cells (Donze et
al. 1996; Tang et al. 1997). Thus
strategies can be pursued to construct novel transcriptional activators that
will recognize the altered binding sites in the wildelta-type delta-globin gene
(expressed at a low level) to attempt to increase the level of expression in
erythroid cells. Success in this
endeavor may lead to new strategies for therapy for patients with sickle cell
disease.
5.8. Recruitment to fetal expression of the
gamma-globin gene in higher primates
Increased
concentrations of Hb F (alpha2gamma2) in erythrocytes will ameliorate the
symptoms of both sickle cell disease and thalassemia (see Chapters 14 by G.
Stamatoyannopoulos, 25 by R. Nagel and M. Steinberg, and 36 by G. Rodgers and
M. Steinberg). Thus considerable
effort has gone into understanding the fetal specific expression of the human
gamma-globin genes and mutations that cause its continued expression in adult
life, leading to the syndrome of hereditary persistence of fetal hemoglobin
(see Chapters 13 by B. Wood and 14 by G. Stamatoyannopoulos). This section will provide an
evolutionary context for these studies, emphasizing the efficacy of
phylogenetic comparisons in generating hypotheses about cis-regulatory elements.
As
discussed in Section 5.3, most eutherian mammals (including prosimians) express
the gamma-globin gene only in embryonic red cells, but anthropoid primates
(monkeys, apes and humans) express it abundantly in fetal red cells. The appearance of this new pattern of fetal
expression coincides with the duplication of the gamma-globin genes in an
ancestral simian, which leads to the hypothesis that the duplication allowed
the changes that caused the fetal recruitment (Fitch et al. 1991; Hayasaka et
al. 1993). Concomitantly,
expression of the beta-globin gene is delayed until just before birth in the
anthropoid primates. In other
mammals, beta-globin gene expression initiates and predominates in the fetal
liver, arguing that fetal expression of the beta-globin gene is the ancestral
state. Note that these changes in
pattern of expression in anthropoid primates are in the definitive erythroid
cell lineage (derived from fetal liver and adult bone marrow), but they do not
necessarily require changes in expression in the primitive erythroid cell
lineage derived from embryonic yolk sac cells (see Chapter 2 by E. Dzierzak).
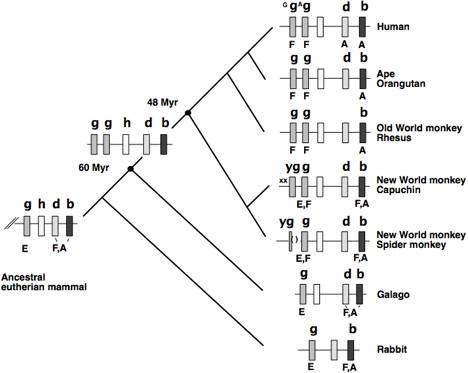
Fig. 12. Recruitment of the gamma-globin gene to
fetal expression in simian primates primates. The portion of the beta-globin gene clusters from gamma-globin
through beta-globin genes is shown, along with the deduced gene duplication in
the ancestor to anthropoid primates. Conventions and abbreviations are as in
other figures. g=gamma, h=eta, d=delta, b=beta, y=psi.
Further
analysis reveals some heterogeneity in the timing of expression of gamma-globin
genes in anthropoid primates (Fig. 12).
As in humans, two gamma-globin genes are found in apes and Old World
monkeys (Slightom et al. 1987; Slightom et al. 1988), and presumably these are
actively expressed in fetal erythroid cells. Duplicated gamma-globin genes are also found in New World
monkeys, but there is a tendency to inactivate one of the copies, either by
partial deletion or by mutations that decrease expression (reviewed in Chiu et
al. 1997). Fetal erythrocytes from
three species of New World monkeys contain both gamma- and beta-globins
(Johnson et al. 1996). This shows
that fetal recruitment of gamma-globin gene expression predates the divergence
between New World and Old World monkeys (about 48 Myr ago) but follows the
simian-prosimian split (about 60 Myr ago). Interestingly, the switch in synthesis from gamma-globin to
beta-globin appears to occur earlier in New World monkeys than in Old World
monkeys and apes. Thus the
gamma-globin genes in New World monkeys may represent an intermediate state in
which the change in timing of expression has not completely shifted to that
seen in humans.
The
cis-elements close to
the gamma-globin gene are key determinants of fetal versus embryonic
expression, as shown by the fact that in otherwise identical constructs, a
prosimian gamma-globin gene is expressed embryonically in transgenic mice,
whereas a human gamma-globin gene is expressed fetally (TomHon et al.
1997). One would like to identify
alterations in the regulatory regions of anthropoid gamma-globin genes that are
associated with this change in stage-specificity, i.e. sequences that are
conserved in anthropoid primates but are different in prosimians and
non-primate mammals. Blocks of
aligned sequences with such properties are called differential phylogenetic
footprints (Gumucio et al. 1994) . This approach led to the identification of a
stage selector element (SSE) in the human gamma-globin gene promoter (Fig.
8). This is a binding site for a
factor called the stage-selector protein, or SSP, that has been implicated in
the differential expression of gamma-globin and beta-globin genes (Jane et al.
1992). SSP is a heterodimer (Jane
et al. 1995) between CP2 (Lim et al. 1992) and another protein. Interestingly, the NFE4 protein
implicated in the stage-specific expression of the chicken beta-globin gene
(Foley and Engel 1992) also contains CP2 as part of a heterodimer (Jane et al.
1995). Further analysis has
revealed an additional DNA sequence that binds several proteins implicated in
fetal silencing of the gamma-globin gene (Gumucio et al. 1994).
The
enhancers located 3' to the Agamma- and beta-globin genes may also be involved
in the changes in timing of expression in anthropoid primates. The 3' Agamma-globin gene enhancer is not conserved in prosimians or
nonprimate mammals (Fig. 9 and Fig. 10); in fact two of the GATA1-binding sites
(Puruker et al. 1990) were brought in via a transposable element (Fig. 10).
Thus the presence of this enhancer correlates with the fetal recruitment of
gamma-globin gene expression. The
3'beta-globin gene enhancer is characterized by several GATA sites (Wall et al.
1988), but these are not conserved in non-anthropoid mammals (Fig. 10). Experiments in transgenic mice revealed
effects of this enhancer primarily on developmental timing (Trudel and
Costantini 1987; Antoniou et al. 1988; Perezeta-Stable and Costantini
1990). It is possible that the 3'
enhancers, as well as promoter regions, contain cis-regulatory elements important in the
fetal recruitment of gamma-globin genes as well as the delay in expression of
the beta-globin gene. Increased
understanding of the role and mechanism of action of proteins implicated in
activation of gamma-globin gene expression (Fig. 8), such as SSP, gPE (Lloyd et
al. 1994), GATA1 (Martin et al. 1989; Puruker et al. 1990; McDonagh et al.
1991), and proteins binding to the CACC and CCAAT motifs, could lead to novel
strategies to increase gamma-globin production in sickle cell disease and
thalassemia.
Acknowledgments
I
thank Webb Miller for producing Figs. 9 and 10. Work in this laboratory is support by the National
Institutes of Health, grant RO1DK27635, and the National Library of Medicine,
grants RO1LM05110 and RO1LM05773.
References
Andersson, C. R., E. O. Jensen, D. J.
Llewellyn, E. S. Dennis and W. J. Peacock (1996) A new hemoglobin gene from
soybean: A role for hemoglobin in all plants. Proc. Natl. Acad. Sci., USA
93:
Antequera, F. B., A. (1993) Number of CpG
islands and genes in human and mouse. Proc. Natl. Acad. Sci., USA 90: 11995-11999.
Antoniou, M., E. deBoer, G. Habets and F.
Grosveld (1988) The human beta-globin gene contains multiple regulatory
regions: Identification of one promoter and two downstream enhancers. EMBO
J. 7: 377-384.
Aparicio, S., A. Morrison, A. Gould, J.
Gilthorpe, C. Chaudhuri, P. Rigby, R. Krumlauf and S. Brenner (1995) Detecting
conserved regulatory elements with the model genome of the Japanese puffer
fish, Fugu rubripes. Proc.
Natl. Acad. Sci., USA 92:
1684-1688.
Appleby, C. A. (1984) Leghemoglobin and
Rhizobium respiration. Ann. Rev. Plant Physiol. 35: 443-478.
Barnhart, K., C. Kim, S. Banerji and M.
Sheffery (1988) Identification and characterization of multiple erythroid cell
proteins that interact with the promoter of the murine alpha-globin gene. Molecular
and Cellular Biology 9:
3215-3226.
Baron, M. H. (1997) Transcriptional
control of globin gene switching during vertebrate development. Biochim
Biophys Acta 1351:
51-72.
Bernardi, G., B. Olofsson, J. Filipski,
M. Zerial, J. Salinas, G. Cuny, M. Meunier-Rotival and F. Rodier (1985) The
mosaic genome of warm-blooded vertebrates. Science 228: 953-958.
Bird, A., M. Taggart, R. Nicholls and D.
Higgs (1987) Non-methylated CpGAMMA-rich islands at the human alpha-globin
locus: implications for evolution of the alpha-globin pseudogene. EMBO J.
6: 999-1004.
Bodine, D. and T. Ley (1987) An enhancer
element lies 3' to the human A gamma globin gene. EMBO J 6: 2997-3004.
Bonaventura, C., Ferruzzi, G., Tesh, S.
and Stevens, R. D. (1999). Effects of S-nitrosation on oxygen binding by normal
and sickle cell hemoglobin. J Biol Chem 274: 24742-24748.
Brisson, N. and D. P. Verma (1982)
Soybean leghemoglobin gene family: normal, pseudo, and truncated genes. Proc.
Natl. Acad. Sci., U. S. A. 79:
4055-4059.
Bulger, M., von Doorninck, J. H., Saitoh,
N., Telling, A., Farrell, C., Bender, M. A., Felsenfeld, G., Axel, R. and
Groudine, M. (1999). Conservation of sequence and structure flanking the mouse
and human beta-globin loci: the beta-globin genes are embedded within an array
of odorant receptor genes. Proc Natl Acad Sci U S A 96: 5129-34.
Bunn, H. F. and B. G. Forget (1986)
Animal Hemoglobins. Hemoglobin: Molecular, Genetic and Clinical Aspects
ed. (W. B. Saunders Co.,
Philadelphia) 126-167.
Calza, R. E., L. A. Eckhardt, T. Giudice
and C. L. Schildkraut (1984) Changes in gene position are accompanied by a
change in time of replication. Cell 36: 689-696.
Caterina, J. J., D. J. Ciavatta, D.
Donze, R. R. Behringer and T. M. Townes (1994) Multiple elements in human
beta-globin locus control region 5' HS2 are involved in enhancer activity and
position-independent transgene expression. Nucl. Acids Res. 22: 1006-1011.
Chan, F., J. Robinson, A. Brownlie, R.
Shivdasani, A. Donovan, C. Brugnara, J. Kim, B. Lau, H. Witkowska and L. Zon
(1997) Characterization of adult alpha- and beta-globin genes in the zebrafish.
Blood 89:
688-700.
Cheng, J., L. Raid and R. C. Hardison
(1986) Isolation and nucleotide sequence of the rabbit globin gene cluster
psuedoZETA-alpha1-psuedoA. J. Biol. Chem. 261: 839-48.
Cheng, J., L. Raid and R. C. Hardison
(1987) Block duplications of a z-z-a-t gene set in the rabbit alphalike globin
gene cluster. J. Biol. Chem. 262: 5414-5421.
Chiu, C.-H., H. Schneider, J. L.
Slightom, D. L. Gumucio and M. Goodman (1997) Dynamics of regulatory evolution
in primate beta-globin gene clusters:
cis-mediated
acquisition of simian g fetal expression patterns. Gene 205: 47-57.
Clark, D., M. Reitman, V. Studitsky, J.
Chung, H. Westphal, E. Lee and F. G. (1993) Chromatin structure of
transcriptionally active genes. Cold Spring Harb. Symp. Quant. Biol. 58: 1-6.
Cohen, R. B., M. Sheffery and C. G. Kim
(1986) Partial purification of a nuclear protein that binds to the CCAAT box of
the mouse a1-globin gene. Mol. Cell. Biol. 6: 821-832.
Collins, F. S. and S. M. Weissman (1984)
The molecular genetics of human hemoglobin. Prog. Nucl. Acids Res. &
Mol. Biol. 31:
315-462.
Cooper, S., R. Murphy, G. Dolman, D.
Hussey and R. Hope (1996) A molecular and evolutionary study of the beta-globin
gene family of the Australian marsupial Sminthopsis crassicaudata. Mol Biol
Evol 13:
1012-1022.
Couture, M., H. Chamberland, B.
St.-Pierre, J. Lafontaine and M. Guertin (1994) Nuclear genes encoding
chloroplast hemoglobins in the unicellular green alga Chlamydomonas
eugametos. Mol. Gen.
Genet. 243:
185-197.
Couture, M. and M. Guertin (1996)
Purification and spectroscopic characterization of a recombinant chloroplastic
hemoglobin from the green unicellular alga Chlamydomonas eugametos. Eur. J. Biochem. 242: 779-787.
Craddock, C. F., P. Vyas, J. A. Sharpe,
H. Ayyub, W. G. Wood and D. R. Higgs (1995) Contrasting effects of alpha and
beta globin regulatory elements on chromatin structure may be related to their
different chromosomal environments. EMBO J. 14: 1718-1726.
Cramm, R., R. A. Siddiqui and B.
Friedrich (1994) Primary structure and evidence for a physiological function of
the flavohemoprotien of Alcaligenes eutrophus. J. Biol. Chem. 269: 7349-7354.
Crawford, M. J. and Goldberg, D. E.
(1998). Role for the Salmonella flavohemoglobin in protection from nitric
oxide. J Biol Chem 273: 12543-7.
deBoer, E., M. Antoniou, V. Mignotte, L.
Wall and F. Grosveld (1988) The human beta-globin promoter; nuclear protein factors and erythroid
specific induction of transcription. EMBO J. 7: 4203-4212.
Deisseroth, A., A. Nienhuis, P. Turner,
R. Velez, W. F. Anderson, F. H. Ruddle, J. Lawrence, R. P. Creagan and R. S.
Kucherlapati (1977) Localization of the human alpha globin structural gene to
chromosome 16 in somatic cell hybrids by molecular hybridization assay. Cell
12: 205-218.
Deisseroth, A., A. W. Nienhuis, J.
Lawrence, R. E. Giles, P. Turner and F. H. Ruddle (1978) Chromosomal
localization of the human beta globin gene to human chromosome 11 in somatic
cell hybrids. Proc. Nat. Acad. Sci., USA 75: 1456-1460.
Deisseroth, A., R. Velez and A. W.
Nienhuis (1976) Hemoglobin synthesis in somatic cell hybrids: independent
segregation of the human alpha- and beta-globin genes. Science 191: 1262-1263.
Delvoye, N. L., N. M. Destroismaisons and
L. A. Wall (1993) Activation of the beta-globin gene promoter by the locus
control region correlates with binding of a novel factor to the CCAAT bos in
murine erythroleukemia cells but not in K562 cells. Mol. Cell. Biol. 13: 6969-6983.
Dhar, V., D. Mager, A. Iqbal and C. L.
Schildkraut (1988) The co-ordinate replication of the human beta-globin gene
domain reflects its transcriptional activity and nuclease hypersensitivity. Mol.
Cell. Biol. 8:
4958-4965.
Dhar, V., A. Nandi, C. L. Schildkraut and
A. I. Skoultchi (1990) Erythroid-specific nucleasepsilon-hypersensitive sites
flanking the human beta-globin gene cluster. Mol. Cell. Biol. 10: 4324-4333.
Dickerson, R. E. and I. Geis (1983) Hemoglobin:
Structure, Function, Evolution and Pathology. Menlo Park, CA, The
Benjamin/Cummings Publishing Co., Inc.
Dikshit, R. P., K. L. Dikshit, Y. Liu and
D. A. Webster (1992) The bacterial hemoglobin from Vitreoscilla can support aerobic growth of Escherichia
coli lacking terminal
oxidases. Arch. Biochem. Biophys. 293: 241-245.
Dixon, B., B. Walker, W. Kimmins and B.
Pohajdak (1992) A nematode hemoglobin gene contains an intron previously
thought to be unique to plants. J. Mol. Evol. 35: 131-136.
Donze, D., P. Jeancake and T. Townes
(1996) Activation of delta-globin gene expression by erythroid
Krupplepsilon-like factor: a potential approach for gene therapy of sickle cell
disease. Blood 88:
4051-4057.
Donze, D., T. M. Townes and J. J. Bieker
(1995) Role of erythroid kruppel-like factor in human gamma- to beta-globin
gene switching. J. Biol. Chem. 270: 1955-1959.
Durner, J., Gow, A. J., Stamler, J. S.
and Glazebrook, J. (1999). Ancient origins of nitric oxide signaling in
biological systems. Proc Natl Acad Sci U S A 96: 14206-7.
Efstratiadis, A., J. W. Posakony, T.
Maniatis, R. M. Lawn, C. O'Connell, R. A. Spritz, J. K. DeRiel, B. G. Forget,
S. M. Weissman, J. L. Slightom, A. E. Blechl, O. Smithies, F. E. Baralle, C. C.
Shoulders and N. J. Proudfoot (1980) The structure and evolution of the human
beta-globin gene family. Cell 21: 653-668.
Elder, J. T., W. C. Forrester, C.
Thompson, D. Mager, P. Henthorn, M. Peretz, T. Papayannopoulou and M. Groudine
(1990) Translocation of an erythroid-specific hypersensitive site in
deletion-type hereditary persistence of fetal hemoglobin. Mol. Cell. Biol.
10: 1382-1389.
Elnitski, L., W. Miller and R. Hardison
(1997) Conserved E boxes function as part of the enhancer in hypersensitive
site 2 of the beta-globin locus control region: Role of basic helix-loop-helix
proteins. J. Biol. Chem. 272:
369-378.
Ermler, U., R. A. Siddiqui, R. Cramm and
B. Friedrich (1995) Crystal structure of the flavohemoglobin from Alcaligenes
eutrophus at 1.75
Angstrom resolution. EMBO J. 14: 6067-6077.
Evans, T., G. Felsenfeld and M. Reitman
(1990) Control of globin gene transcription. Annu. Rev. Cell Biol. 6: 95-124.
Feingold, E. A. and Forget, B. G. (1989).
The breakpoint of a large deletion causing hereditary persistence of fetal
hemoglobin occurs within an erythroid DNA domain remote from the beta-globin
gene cluster. Blood 74: 2178-86.
Feingold, E. A., Penny, L. A., Nienhuis,
A. W. and Forget, B. G. (1999). An olfactory receptor gene is located in the
extended human beta-globin gene cluster and is expressed in erythroid cells. Genomics
61: 15-23.
Feng, D. F., G. Cho and R. F. Doolittle
(1997) Determining divergence times with a protein clock: update and
reevaluation. Proc. Natl. Acad. Sci., U S A 94: 13028-13033.
Fischel-Ghodsian, N., R. D. Nicholls and
D. R. Higgs (1987) Unusual features of CpGAMMA-rich (HTF) islands in the human
alpha-globin complex: association
with nonfunctional pseudogenes and presence within the 3' portion of the z
genes. Nucl. Acids Res. 15:
9215-9225.
Fitch, D. H., W. J. Bailey, D. A. Tagle,
M. Goodman, L. Sieu and J. L. Slightom (1991) Duplication of the gamma-globin
gene mediated by L1 long interspersed repetitive elements in an early ancestor
of simian primates. Proc. Natl. Acad. Sci. U. S. A. 88: 7396-7400.
Flint, J., A. M. Taylor and J. B. Clegg
(1988) Structure and evolution of the horse zeta globin locus. J. Mol. Biol.
199: 427-437.
Flint, J., K. Thomas, G. Micklem, H.
Raynham, K. Clark, N. A. Doggett, A. King and D. R. Higgs (1997) The
relationship between chromosome structure and function at a human telomeric
region. Nature Genetics 15:
252-257.
Foley, K. P. and J. D. Engel (1992)
Individual stage selector element mutations lead to reciprocal changes in beta-
vs. epsilon-globin gene transcription: genetic confirmation of promoter
competition during globin gene switching. Genes & Devel. 6: 730-744.
Forrester, W., S. Takegawa, T.
Papayannopoulou, G. Stamatoyannopoulos and M. Groudine (1987) Evidence for a
locus activating region: The formation of developmentally stable hypersensitive
sites in globin-expressing hybrids. Nucl. Acids Res. 15: 10159-10177.
Forrester, W. C., E. Epner, M. C.
Driscoll, T. Enver, M. Brice, T. Papayannopoulou and M. Groudine (1990) A
deletion of the human beta-globin locus activation region causes a major
alteration in chromatin structure and replication across the entire beta-globin
locus. Genes & Devel. 4:
1637-1649.
Fraser, P., J. Hurst, P. Collis and F.
Grosveld (1990) DNase I hypersensitive sites 1, 2 and 3 of the human
beta-globin dominant control region direct position-independent expression. Nucl.
Acids Res. 18:
3503-3508.
Fritsch, E., R. Lawn and T. Maniatis
(1980) Molecular cloning and characterization of the human beta-like globin
gene cluster. Cell 19:
959-972.
Gardner, P. R., Gardner, A. M., Martin,
L. A. and Salzman, A. L. (1998). Nitric oxide dioxygenase: an enzymic function
for flavohemoglobin. Proc Natl Acad Sci U S A 95: 10378-83.
Gilbert, W. (1978) Why genes in pieces? Nature
271: 501.
Gladwin, M. T., Schechter, A. N.,
Shelhamer, J. H., Pannell, L. K., Conway, D. A., Hrinczenko, B. W., Nichols, J.
S., Peasepsilon-Fye, M. E., Noguchi, C. T., Rodgers, G. P. and Ognibene, F. P.
(1999). Inhaled nitric oxide augments nitric oxide transport on sickle cell
hemoglobin without affecting oxygen affinity. J Clin Invest 104: 937-45.
Go, M. (1981) Correlation of DNA exonic
regions with protein structural units in haemoglobin. Nature 291: 90-92.
Gong, Q.-H. and A. Dean (1993)
Enhancer-dependent transcripion of the epsilon-globin promoter requires
promoter-bound GATA-1 and enhancer-bound AP-1/NF-E2. Mol. Cell. Biol. 13: 911-917.
Goodman, M., J. Czelusniak, B. Koop, D.
Tagle and J. Slightom (1987) Globins:
A case study in molecular phylogeny. Cold Spring Harbor Symp. Quant.
Biol. 52:
875-890.
Goodman, M., B. F. Koop, J. Czelusniak,
M. L. Weiss and J. L. Sightom (1984) The eta-globin gene: its long evolutionary
history in the beta-globin gene families of mammals. J. Mol. Biol. 180: 803-824.
Gottgens, B., Barton, L. M., Gilbert, J.
G., Bench, A. J., Sanchez, M. J., Bahn, S., Mistry, S., Grafham, D., McMurray,
A., Vaudin, M., Amaya, E., Bentley, D. R. and Green, A. R. (2000). Analysis of
vertebrate SCL loci
identifies conserved enhancers. Nat Biotechnol 18: 181-6.
Gourdon, G., J. A. Sharpe, D. R. Higgs
and W. G. Wood (1995) The mouse alpha-globin locus regulatory element. Blood
86: 766-775.
Gow, A. J. and Stamler, J. S. (1998). Reactions
between nitric oxide and haemoglobin under physiological conditions. Nature
391: 169-73.
Grosveld, F., M. Antoniou, M. Berry, E.
de Boer, N. Dillon, J. Ellis, P. Fraser, O. Hanscombe, J. Hurst, A. Imam, M.
Lindenbaum, S. Philipsen, S. Pruzina, J. Strouboulis, S. Raguzeta-Bolognesi and
D. Talbot (1993) The regulation of human globin gene switching. Philos.
Trans. R. Soc. Lond. 339:
183-191.
Grosveld, F., G. B. van Assendelft, D.
Greaves and G. Kollias (1987) Position-independent, higeta-level expression of
the human beta-globin gene in transgenic mice. Cell 51: 975-985.
Groudine, J., T. Kohwi-Shigematsu, R.
Gelinas, G. Stamatoyannopoylos and T. Papyannopoulou (1983) Human fetal to
adult hemoglobin switching:
Changes in chromatin structure of the beta-globin gene locus. Proc.
Natl. Acad. Sci., USA 80:
7551-7555.
Gumucio, D., D. Shelton, W. Zhu, D.
Millinoff, T. Gray, J. Bock, J. Slightom and M. Goodman (1996) Evolutionary
strategies for the elucidation of cis and trans
factors that regulate the developmental switching programs of the beta-like
globin genes. Mol. Phylog. and Evol. 5: 18-32.
Gumucio, D. L., H. Heilstedt-Williamson,
T. A. Gray, S. A. Tarle, D. A. Shelton, D. Tagle, J. Slightom, M. Goodman and
F. S. Collins (1992) Phylogenetic footprinting reveals a nuclear protein which
binds to silencer sequences in the human g and e globin genes. Mol. Cell.
Biol. 12:
4919-4929.
Gumucio, D. L., D. A. Shelton, W. J.
Bailey, J. L. Slightom and M. Goodman (1993) Phylogenetic footprinting reveals
unexpected complexity in trans factor binding upstream from the e-globin gene. Proc.
Natl. Acad. Sci., USA 90:
6018-6022.
Gumucio, D. L., D. A. Shelton, K.
Blanchardelta-McQuate, T. A. Gray, S. A. Tarle, H. Heilstedt-Williamson, J.
Slightom, F. S. Collins and M. Goodman (1994) Differential phylogenetic
footprinting as a means to identify base changes responsible for recruitment of
the anthropoid g gene to a fetal expression pattern. J. Biol. Chem. 269: 15371-15380.
Hardies, S. C., M. H. Edgell and C. A.
Hutchison III (1984) Evolution of the mammalian b-globin gene cluster. J.
Biol. Chem. 259:
3748-3756.
Hardison, R. (1998) Hemoglobins from
bacteria to man: evolution of different patterns of gene expression. J.
Experimental Biology 201:
1099-1117.
Hardison, R., K.-M. Chao, S. Schwartz, N.
Stojanovic, M. Ganetsky and W. Miller (1994) Globin gene server: A prototype EPSILON-mail database
server featuring extensive multiple alignments and data compilation. Genomics
21: 344-353.
Hardison, R., D. Krane, D. Vandenbergh,
J.-F. Cheng, J. Mansberger, J. Taddie, S. Schwartz, X. Huang and W. Miller
(1991) Sequence and comparative analysis of the rabbit a-like globin gene
cluster reveals a rapid mode of evolution in a G+C rich region of mammalian
genomes. J. Mol. Biol. 222:
233-249.
Hardison, R. and W. Miller (1993) Use of
long sequence alignments to study the evolution and regulation of mammalian
globin gene clusters. Mol. Biol. Evol. 10: 73-102.
Hardison, R., J. Oeltjen and W. Miller
(1997a) Long human-mouse sequence alignments reveal novel regulatory
elements: A reason to sequence the
mouse genome. Genome Research 7: 959-966.
Hardison, R., J. L. Slightom, D. L.
Gumucio, M. Goodman, N. Stojanovic and W. Miller (1997b) Locus control regions
of mammalian beta-globin gene clusters: Combining phylogenetic analyses and
experimental results to gain functional insights. Gene 205: 73-94.
Hardison, R., J. Xu, J. Jackson, J.
Mansberger, O. Selifonova, B. Grotch, J. Biesecker, H. Petrykowska and W.
Miller (1993) Comparative analysis of the locus control region of the rabbit
beta-like globin gene cluster: HS3
increases transient expression of an embryonic e-globin gene. Nucl. Acids
Res. 21:
1265-1272.
Hardison, R. C. (1983) The nucleotide
sequence of the rabbit embryonic globin gene b4. J. Biol. Chem. 258: 8739-8744.
Hardison, R. C. (1984) Comparison of the
beta-like globin gene families of rabbits and humans indicates that the gene
cluster 5' epsilon-gamma-delta-beta 3' predates the mammalian radiation. Mol.
Biol. Evol. 1:
390-410.
Hardison, R. C. (1991) Evolution of
globin gene families. Evolution at the Molecular Level R. K. Selander,
T. S. Whittam and A. G. Clark, ed.
(Sinauer Associates, Inc., Sunderland, MA) 272-289.
Hardison, R. C. (1996) A brief history of
hemoglobins: Plant, animal, protist and bacteria. Proc. Natl. Acad. Sci.,
USA 93:
5675-5679.
Hardison, R. C. and R. Gelinas (1986)
Assignment of orthologous relationships among mammalian a-globin genes by
examining flanking regions reveals a rapid rate of evolution. Molecular
Biology of Evolution 3:
243-261.
Hardison, R. C. and J. B. Margot (1984)
Rabbit globin pseudogene pseudobeta2 is a hybrid of delta- and beta-globin gene
sequences. Mol. Biol. Evol. 1: 302-316.
Hausladen, A., Gow, A. J. and Stamler, J.
S. (1998). Nitrosative stress: metabolic pathway involving the flavohemoglobin.
Proc Natl Acad Sci U S A 95: 14100-5.
Hayasaka, K., C. Skinner, M. Goodman and
J. Slightom (1993) The gamma-globin genes and their flanking sequences in
primates: findings with nucleotide sequences of capuchin monkey and tarsier. Genomics
18: 20-28.
Hebbes, T. R., A. L. Clayton, A. W.
Thorne and C. Cranepsilon-Robinson (1994) Core histone hyperacetylation co-maps
with generalized DNase I sensitivity in the chicken beta-globin chromosomal
domain. EMBO J. 13:
1823-1830.
Heller, P. and V. Yakulis (1969) The
distribution of hemoglobin A2. Ann.
N.Y. Acad. Sci. 165:
54-59.
Hess, J., C. Schmid and C. Shen (1984) A
gradient of sequence divergence in the human adult alpha-globin duplication
units. Science 226:
67-70.
Higgs, D., W. Wood, A. Jarman, J. Sharpe,
J. Lida, I. M. Pretorious and H. Ayyub (1990) A major positive regulatory
region located far upstream of the human alpha-globin gene locus. Genes
& Devel. 4:
1588-1601.
Hooft van Huijsduijnen, R., X. Y. Li, D.
Black, H. Matthes, C. Benoist and D. Mathis (1990) Co-evolution from yeast to
mouse: cDNA cloning of the two
NF-Y (CP-1/CBF) subunits. The EMBO J. 9: 3119-3127.
Hosbach, H. A., T. Wyler and R. Weber
(1983) The Xenopus laevis
globin gene family: Chromosomal arrangement and gene structure. Cell 32: 45-53.
Hsu, S., J. Marks, J. Shaw, M. Tam, D.
Higgs, C. Shen and C. Shen (1988) Structure and expression of the human theta 1
globin gene. Nature 331:
94-96.
Hug, B. A., A. M. Moon and T. J. Ley
(1992) Structure and function of the murine beta-globin locus control region 5'
HS-3. Nucl. Acids Res. 21:
5771-5778.
Hughes, S., E. Stubblefield, F. Payvar,
J. Engel, J. Dodgson, D. Spector, B. Cordell, R. Schimke and H. Varmus (1979)
Gene localization by chromosome fractionation: globin genes are on at least two
chromosomes and three estrogen-inducible genes are on three chromosomes. Proc
Natl Acad Sci, U S A 76:
1348-1352.
Hyldig-Nielsen, J. J., E. O. Jensen, K.
Paludan, O. Wiborg, R. Garrett, P. Jorgensen and K. A. Marcker (1982) The
primary structures of two leghemoglobin genes from soybean. Nucleic Acids
Res. 10: 689-701.
Jane, S., A. Nienhuis and J. Cunningham
(1995) Hemoglobin switching in man and chicken is mediated by a heteromeric
complex between the ubiquitous transcription factor CP2 and a developmentally
specific protein. EMBO J. 14:
97-105.
Jane, S. M., P. A. Ney, E. F. Vanin, D.
L. Gumucio and A. W. Nienhuis (1992) Identification of a stage selector element
in the human gamma-globin gene promoter that fosters preferential interaction
with the 5' HS2 enhancer when in competition with the beta-promoter. EMBO J.
11: 2961-2969.
Jarman, A., W. Wood, J. Sharpe, G.
Gourdon, H. Ayyub and D. Higgs (1991) Characterization of the major regulatory
element upstream of the human alpha-globin gene cluster. Mol. Cell. Biol.
11: 4679-4689.
Jia, L., Bonaventura, C., Bonaventura, J.
and Stamler, J. S. (1996). S-nitrosohaemoglobin: a dynamic activity of blood
involved in vascular control. Nature 380: 221-6.
Jimenez, G., K. B. Gale and T. Enver
(1992) The mouse beta-globin locus control region: hypersensitive sites 3 and 4. Nucl. Acids Res. 20: 5797-5803.
Johnson, R. M., S. Buck, C.-h. Chiu, H.
Schneider, I. Sampaio, D. A. Gage, T.-L. Shen, M. P. C. Schneider, J. A. Muniz,
D. L. Gumucio and M. Goodman (1996) Fetal globin expression in New World
monkeys. J. Biol. Chem. 271:
14684-14691.
Kim, J., C. Yu, A. Bailey, R. Hardison
and C. Shen (1989) Unique sequence organization and erythroid cell-specific
nuclear factor-binding of mammalian theta 1 globin promoters. Nucleic Acids
Res 17:
5687-5700.
Koop, B. and M. Goodman (1988)
Evolutionary and developmental aspects of two hemoglobin beta-chain genes
(epsilon M and beta M) of opossum. Proc Natl Acad Sci, U S A 85: 3893-3897.
Kosteas, T., Palena, A. and Anagnou, N.
P. (1997). Molecular cloning of the breakpoints of the hereditary persisptence
of fetal hemoglobin type-6 (HPFH-6) deletion and sequence analysis of the novel
juxtaposed region from the 3' end of the beta-globin gene cluster. Hum Genet
100: 441-5.
Kumar, S. and S. B. Hedges (1998) A
molecular timescale for vertebrate evolution. Nature 392: 917-920.
Kumar, S., K. Tamura and M. Nei (1993) MEGA:
Molecular evolutionary genetic analysis, version 1.01. University Park, PA,
The Pennsylvania State University.
Lacy, E. and T. Maniatis (1980)
Nucleotide sequence of the rabbit pseudogene psi beta1. Cell 21: 545-553.
Lam, L. and E. H. Bresnick (1996) A novel
DNA binding protein, HS2NF5, interacts with a functionally important sequence
of the human beta-globin locus control region. J. Biol. Chem. 271: 32421-32429.
Lauer, J., C. Shen and T. Maniatis (1980)
The Chromosomal Arrangement of Human alpha-Like Globin Genes: Sequence Homology
and alpha- Globin Gene Deletions. Cell 20: 119-130.
Leder, A., L. Weir and P. Leder (1985)
Characterization, expression and evolution of the mouse embryonic zeta-globin
gene. Molecular and Cellular Biology 5: 1025-1033.
Lee, J. S., G. G. Brown and D. P. S.
Verma (1983) Chromosomal arrangement of leghemoglobin genes in soybeans. Nucleic
Acids Res. 11:
5541-5552.
Leung, S., E. Whitelaw and N. Proudfoot
(1989) Transcriptional and translational analysis of the human theta globin gene.
Nucleic Acids Res 17:
8283-8300.
Li, Q., P. A. Powers and O. Smithies
(1985) Nucleotide sequence of 16-kilobase pairs of DNA 5' to the human
epsilon-globin gene. J. Biol. Chem. 260: 14901-14910.
Li, Q., B. Zhou, P. Powers, T. Enver and
G. Stamatoyannopoulos (1991) Primary structure of the goat beta-globin locus
control region. Genomics 9:
488-499.
Lim, L. C., S. L. Swendeman and M.
Sheffery (1992) Molecular cloning of the alpha-globin transcription factor CP2.
Mol. Cell. Biol. 12:
828-835.
Liu, L., Zeng, M., Hausladen, A.,
Heitman, J. and Stamler, J. S. (2000). Protection from nitrosative stress by
yeast flavohemoglobin. Proc Natl Acad Sci U S A , 97: 4672-4676.
Lloyd, J. A., S. S. Case, E. Ponce and J.
B. Lingrel (1994) Positive transcriptional regulation of the human gamma-globin
gene: gammaPE is a novel nuclear factor with multiple binding sites near the
gene. J. Biol. Chem. 269:
26-34.
Macleod, K. and M. Plumb (1991)
Derepression of mouse beta-major-globin gene transcription during erythroid
differentiation. Mol. Cell. Biol. 11: 4324-4332.
Margot, J. B., G. W. Demers and R. C.
Hardison (1989) Complete nucleotide sequence of the rabbit beta-like globin
gene cluster: Analysis of intergenic sequences and comparison with the human
beta-like globin gene cluster. J. Mol. Biol. 205: 15-40.
Martin, D. I. K., S.-F. Tsai and S. H.
Orkin (1989) Increased gamma-globin expression in a nondeletion HPFH mediated
by an erythroid-specific DNA binding factor. Nature 338: 435-438.
Martin, S. L., K. A. Vincent and A. C. Wilson
(1983) Rise and fall of the delta globin gene. J. Mol. Biol. 164: 513-528.
Matsuo, K., O. Clay, T. Takahashi, J.
Silke and W. Schaffner (1993) Evidence for erosion of mouse CpG islands during
mammalian evolution. Somatic Cell and Molecular Genetics 19: 543-555.
McDonagh, K. T., H. J. Lin, C. H. Lowrey,
D. M. Bodine and A. W. Nienhuis (1991) The upstream region of the human
gamma-globin gene promoter: Identification and functional analysis of nuclear
protein binding sites. J. Biol. Chem. 266: 11965-11974.
Miller, I. J. and J. J. Bieker (1993) A
novel, erythroid cell-specific murine transcription factor that binds to the
CACCC element and is related to the Kruppel family of nuclear factors. Mol. Cell.
Biol. 13:
2776-2786.
Miller, W., M. Boguski, B. Raghavachari,
Z. Zhang and R. C. Hardison (1994) Constructing aligned sequence blocks. J.
Computational Biology 1:
51-64.
Minning, D. M., Gow, A. J., Bonaventura,
J., Braun, R., Dewhirst, M., Goldberg, D. E. and Stamler, J. S. (1999). Ascaris haemoglobin is a nitric
oxidepsilon-activated 'deoxygenase'. Nature 401: 497-502.
Moon, A. M. and T. J. Ley (1990)
Conservation of the primary structure, organization, and function of the human
and mouse beta-globin locus-activating regions. Proc. Natl. Acad. Sci., USA
87: 7693-7697.
Morley, B. J., C. A. Abbott, J. A.
Sharpe, J. Lida, P. S. Chan-Thomas and W. G. Wood (1992) A single beta-globin
locus control region element (5' hypersensitive site 2) is sufficient for
developmental regulation of human globin genes in transgenic mice. Mol.
Cell. Biol. 12:
2057-2066.
Motamed, K., C. Bastiani, Q. Zhang, A.
Bailey and C.-K. J. Shen (1993) CACC box and enhancer response of human
embryonic epsilon globin promoter. Gene 123: 235-240.
Motohashi, H., J. A. Shavit, K. Igarashi,
M. Yamamoto and J. D. Engel (1997) The world according to Maf. Nucleic Acids
Res. 25:
2953-2959.
Nagel, R. L. (1999). Can we just say NO
to sickle cell anemia? J Clin Invest 104: 847-8.
Nagel, R., R. Bookchin, J. Johnson, D.
Labie, H. Wajcman, W. Isaac-Sodeye, G. Honig, G. Schiliro, J. Crookston and K.
Matsutomo (1979) Structural bases of the inhibitory effects of hemoglobin F and
hemoglobin A2 on the polymerization of hemoglobin S. Proc Natl Acad Sci, U S
A 76: 670-672.
Ney, P., B. Sorrentino, K. McDonagh and
A. Nienhuis (1990) Tandem AP-1-binding sites within the human beta-globin
dominant control region function as an inducible enhancer in erythroid cells. Genes
& Devel. 4:
993-1006.
Nishioka, Y. and P. Leder (1979) The
complete sequence of a chromosomal mouse alpha-globin gene reveals elements
conserved throughout vertebrate evolution. Cell 18: 875-882.
Nuez, B., D. Michalovich, A. Bygrave, R.
Ploemacher and F. Grosveld (1995) Defective haematopoiesis in fetal liver
resulting from inactivation of the EKLF gene. Nature 375: 316-318.
Orkin, S. H. (1995) Transcription factors
and hematopoietic development. J. Biol. Chem. 270: 4955-4958.
Perez-Stable, C. and F. Costantini (1990)
Role of fetal Ggamma-globin promoter elements and the adult beta-globin 3'
enhancer in the stage-specific expression of globin genes. Mol. Cell. Biol.
10: 1116-1125.
Perkins, A. C., A. H. Sharpe and S. H.
Orkin (1995) Lethal beta-thalassaemia in mice lacking the erythroid
CACCC-transcription factor EKLF. Nature 375: 318-322.
Peschle, C., F. Mavilio, A. Care, G.
Migliaccio, A. R. Migliaccio, G. Salvo, P. Samoggia, S. Petti, R. Guerriero, M.
Marinucci and e. al. (1985) Hemoglobin switching in human embryos: asynchrony
of zeta-alpha and epsilon-gamma-globin switches in primitive and definite erythropoietic
lineage. Nature 313:
235-238.
Peters, B., N. Merezhinskaya, J. F. X.
Diffley and C. T. Noguchi (1993) Protein-DNA interactions in the epsilon-globin
gene silencer. J. Biol. Chem. 268: 3430-3437.
Poillon, W., B. Kim, G. Rodgers, C.
Noguchi and A. Schechter (1993) Sparing effect of hemoglobin F and hemoglobin
A2 on the polymerization of hemoglobin S at physiologic ligand saturations. Proc
Natl Acad Sci, U S A 90:
5039-5043.
Pondel, M., S. Murphy, L. Pearson, C.
Craddock and N. Proudfoot (1995) Sp1 functions in a chromatin-dependent manner
to augment human alpha-globin promoter activity. Proc Natl Acad Sci U S A
92: 7237-7241.
Pondel, M. D., M. George and N. J.
Proudfoot (1992) The LCR-like alpha-globin positive regulatory element
functions as an enhancer in transiently transfected cells during erythroid
differentiation. Nucl. Acids Res. 20: 237-243.
Prioleau, M. N., Nony, P., Simpson, M.
and Felsenfeld, G. (1999). An insulator element and condensed chromatin region
separate the chicken beta-globin locus from an independently regulated
erythroid-specific folate receptor gene. Embo J 18: 4035-48.
Puruker, M., D. Bodine, H. Lin, K.
McDonagh and A. W. Nienhuis (1990) Structure and function of the enhancer 3' to
the human Agamma-globin gene. Nucleic Acids Res. 18: 407-7415.
Reddy, P. M. S., G. Stamatoyannopoulos,
T. Papayannopoulou and C.-K. J. Shen (1994) Genomic footprinting and sequencing
of human beta-globin locus: Tissue
specificity and cell line artifact. J. Biol. Chem. 269: 8287-8295.
Reitman, M., J. A. Grasso, R. Blumentahl
and P. Lewit (1993) Primary sequence, evolution and repetitive elements of the G.
gallus (chicken)
b-globin cluster. Genomics 18: 616-626.
Reitman, M., E. Lee and H. Westphal
(1995) Function of upstream hypersensitive sites of the chicken beta-gobin gene
cluster in mice. Nucl. Acids Res. 23: 1790-1794.
Reitman, M., E. Lee, H. Westphal and G. Felsenfeld
(1990) Site-independent expression of the chicken betaA-globin gene in
transgenic mice. Nature 348:
749-752.
Ren, S., X.-n. Luo and G. Atweh (1993)
The major regulatory element upstream of the alpha-globin gene has classical
and inducible enhancer activity. Blood 81: 1058-1066.
Riggs, A. F. (1991) Aspects of the origin
and evolution of non-vertebrate hemoglobins. Amer. Zool. 31: 535-545.
Riggs, A. F. (1998) Self-association,
cooperativity and supercooperativity of oxygen binding by hemoglobins. J.
Experimental Biol. 201:
1073-1084.
Rohrbaugh, M. L. and R. C. Hardison
(1983) Analysis of rabbit beta-like globin gene transcripts during development.
Journal of Molecular Biology 164: 395-417.
Rombel, I., K.-Y. Hu, Q. Zhang, T.
Papyannopoulou, G. Stamatoyannopoulos and C.-K. J. Shen (1995) Transcriptional
activation of human alpha-globin gene by hypersensitive site-40 enhancers:
function of nuclear factor-binding motifs occupied in erythroid cells. Proc.
Natl. Acad. Sci., USA 92:
6454-6458.
Sawada, I. and C. Schmid (1986) Primate
evolution of the alpha-globin gene cluster and its Alu-like repeats. J. Mol.
Biol. 192:
693-709.
Schirmer, T., W. Bode, R. Huber, W.
Sidler and H. Zuber (1985) X-ray crystallographic structure of the light
harvesting biliprotein C-phycocyanin from the thermophilic cyanobacterium Mastigocladus
laminosus and its
resemblance to globin structures. J. Mol. Biol. 184: 257-277.
Shehee, R., D. D. Loeb, N. B. Adey, F. H.
Burton, N. C. Casavant, P. Cole, C. J. Davies, R. A. McGraw, S. A. Schichman,
D. M. Severynse, C. F. Voliva, F. W. Weyter, G. B. Wisely, M. H. Edgell and C.
A. Hutchison III (1989) Nucleotide sequence of the BALB/c mouse beta-globin
complex. J. Mol. Biol. 205:
41-62.
Shen, S.-h., J. L. Slightom and O.
Smithies (1981) A history of the human fetal globin gene duplication. Cell
26: 191-203.
Sherman, D. R., A. P. Kloek, B. R.
Krishnan, B. Guinn and D. E. Goldberg (1992) Ascaris hemoglobin gene: Plant-like structure
reflects the ancestral globin gene. Proc. Natl. Acad. Sci., USA 89: 11696-11700.
Shewchuk, B. M. and R. C. Hardison (1997)
CpG islands from the alpha-globin gene cluster increase gene expression in an
integration-dependent manner. Mol. Cell. Biol. 17: 5856-5866.
Slightom, J., J. Bock, D. Tagle, D.
Gumucio, M. Goodman, N. Stojanovic, J. Jackson, W. Miller and R. Hardison
(1997) The complete sequences of the galago and rabbit beta-globin locus
control regions: Extended sequence and functional conservation outside the
cores of DNase hypersensitive sites. Genomics 39: 90-94.
Slightom, J., B. Koop, P. Xu and M.
Goodman (1988) Rhesus fetal globin genes: Concerted gene evolution in the
descent of higher primates. J. Biol. Chem. 263: 12427-12438.
Slightom, J., T. W. Theisen, B. Koop and
M. Goodman (1987) Orangutan fetal globin genes: Nucleotide sequences reveal
multiple gene conversions during hominid phylogeny. J. Biol. Chem. 262: 7472-7483.
Spritz, R., J. DeRiel, B. Forget and S.
Weissman (1980) Complete nucleotide sequence of the human delta-globin gene. Cell
21: 639-646.
Spritz, R. and L. Giebel (1988) The
structure and evolution of the spider monkey delta-globin gene. Mol Biol
Evol 5: 21-29.
Stoltzfus, A., D. F. Spencer, M. Zuker,
J. M. Logsdon Jr. and W. F. Doolittle (1994) Testing the exon theory of genes:
The evidence from protein structure. Science 265: 202-207.
Stamler, J. S., Jia, L., Eu, J. P.,
McMahon, T. J., Demchenko, I. T., Bonaventura, J., Gernert, K. and Piantadosi,
C. A. (1997). Blood flow regulation by S-nitrosohemoglobin in the physiological
oxygen gradient. Science 276: 2034-7.
Strauss, E. C., N. C. Andrews, D. R.
Higgs and S. H. Orkin (1992) In vivo footprinting of the human alpha-globin
locus upstream regulatory element by guanine and adenine ligation-mediated
polymerase chain reaction. Mol. Cell. Biol. 12: 2135-2142.
Stuve, L. L. and R. M. Myers (1990) A
directly repeated sequence in the beta-globin promoter regulates transcription
in murine erythroleukemia cells. Mol. Cell. Biol. 10: 972-981.
Tagle, D., J. Slightom, R. Jones and M.
Goodman (1991) Concerted evolution led to high expression of a prosimian
primate delta globin gene locus. J
Biol Chem 266:
7469-7480.
Tagle, D., M. J. Stanhope, D. R.
Siemieniak, P. Benson, M. Goodman and J. L. Slightom (1992) The beta-globin
gene cluster of the prosimian primate Galago crassicaudatus: Nucleotide sequence determination of
the 41-kb cluster and comparative sequence analysis. Genomics 13: 741-760.
Tagle, D. A., B. F. Koop, M. Goodman, J.
Slightom, D. L. Hess and R. T. Jones (1988) Embryonic epsilon and gamma globin
genes of a prosimian primate (Galago crassicaudatus):
Nucleotide and amino acid sequences, developmental regulation and
phylogenetic footprints. J. Mol. Biol. 203: 7469-7480.
Talbot, D., S. Philipsen, P. Fraser and
F. Grosveld (1990) Detailed analysis of the site 3 region of the human
beta-globin dominant control region. EMBO J. 9: 2169-2178.
Tan, H. and J. I. Whitney (1993) Genomic
rearrangement of the alpha-globin gene complex during mammalian evolution. Biochem
Genet 31:
473-484.
Tang, D. C., D. Ebb, R. C. Hardison and
G. P. Rodgers (1997) Restoration of the CCAAT box and insertion of the CACCC
motif activate delta-globin gene expression. Blood 90: 421-427.
Tarricone, C., A. Galizzi, A. Coda, P.
Ascenzi and M. Bolognesi (1997) Unusual structure of the oxygen-binding site in
the dimeric bacterial hemoglobin from Vitreoscilla sp. Structure 5: 497-507.
Tatusov, R., E. Koonin and D. Lipman
(1997) A genomic perspective on protein families. Science 278: 631-637.
Terwilliger, N. B. (1998) Functional
adaptations of oxygen-transport proteins. J. Experimental Biology 201: 1085-1098.
TomHon, C., W. Zhu, D. Millinoff, K.
Hayasaka, J. L. Slightom, M. Goodman and D. L. Gumucio (1997) Evolution of a
fetal expression pattern via cis-changes
near the gamma-globin gene. J. Biol. Chem. 272: 14062-14066.
Townes, T. M., M. C. Fitzgerald and J. B.
Lingrel (1984) Triplication of a four-gene set during evolution of the goat
beta-globin locus produced three gene now expressed differentially during
development. Proc. Natl. Acad. Sci., USA 81: 6589-6593.
Trudel, M. and F. Costantini (1987) A 3'
enhancer contributes to the stage-specific expression of the human beta-globin
gene. Genes & Devel. 1:
954-961.
Tuan, D., W. Solomon, Q. Li and I. M.
London (1985) The beta-like globin gene domain in human erythroid cells. Proc.
Natl. Acad. Sci., USA 82:
6384-6388.
van der Ploeg, L. H. T. and R. A. Flavell
(1980) DNA methylation in the human gamma-delta-beta globin locus in erythroid
and nonerythroid tissues. Cell 19: 947-958.
Wall, L., E. deBoer and F. Grosveld
(1988) The human beta-globin gene 3' enhancer contains multiple binding sites
for an erythroid-specific protein. Genes & Devel. 2: 1089-1100.
Weintraub, H. and M. Groudine (1976)
Chromosomal subunits in active genes have an altered conformation. Science
193: 848-856.
Wernke, S. M. and J. B. Lingrel (1986)
Nucleotide sequences of the goat embryonic alpha globin gene (zeta) and
evolutionary analysis of the complete alpha globin cluster. J. Mol. Biol.
192: 457-471.
Wittenberg, B. A. and J. B. Wittenberg
(1987) Myoglobin-mediated oxygen delivery to mitochondria of isolated cardiac
myocytes. Proc. Natl. Acad. Sci., USA 84: 7503-7507.
Xu, J. and R. C. Hardison (1991)
Localization of the alpha-like globin gene cluster to region q12 of rabbit
chromosome 6 by in situ
hybridization. Genomics 9:
362-365.
Yamauchi, K., H. Tada and I. Usuki (1995)
Structure and evolution of Paramecium hemoglobin genes. Biochim. Biophys. Acta 1264: 53-62.
Yost, S. E., B. Shewchuk and R. Hardison
(1993) Nuclear protein binding sites in a transcriptional control region of the
rabbit alpha-globin gene. Mol. Cell. Biol. 13: 5439-5449.
Yu, C.-Y., K. Motamed, J. Chen, A. D.
Bailey and C.-K. J. Shen (1991) The CACC box upstream of human embryonic
epsilon globin gene binds Sp1 and is a functional promoter element in vitro and in vivo. J. Biol. Chem. 266: 8907-8915.
Zhu, H. and A. F. Riggs (1992) Yeast
flavohemoglobin is an ancient protein related to globins and a reductase
family. Proc. Natl. Acad. Sci., USA 89: 5015-5019.