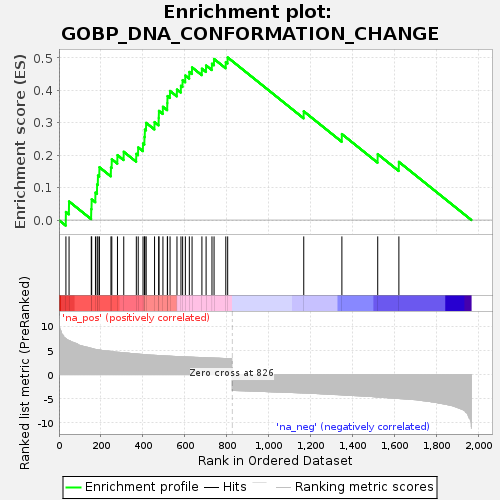
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | H2BC12 | H2B clustered histone 12 [Source:HGNC Symbol;Acc:HGNC:13954] | 33 | 7.423 | 0.0249 | Yes |
| 2 | BLM | BLM RecQ like helicase [Source:HGNC Symbol;Acc:HGNC:1058] | 48 | 7.035 | 0.0574 | Yes |
| 3 | CDKN2A | cyclin dependent kinase inhibitor 2A [Source:HGNC Symbol;Acc:HGNC:1787] | 154 | 5.438 | 0.0337 | Yes |
| 4 | PIF1 | PIF1 5'-to-3' DNA helicase [Source:HGNC Symbol;Acc:HGNC:26220] | 156 | 5.418 | 0.0638 | Yes |
| 5 | H2BC14 | H2B clustered histone 14 [Source:HGNC Symbol;Acc:HGNC:4750] | 174 | 5.208 | 0.0845 | Yes |
| 6 | TTN | titin [Source:HGNC Symbol;Acc:HGNC:12403] | 182 | 5.145 | 0.1099 | Yes |
| 7 | G3BP1 | G3BP stress granule assembly factor 1 [Source:HGNC Symbol;Acc:HGNC:30292] | 186 | 5.119 | 0.1373 | Yes |
| 8 | H2BC7 | H2B clustered histone 7 [Source:HGNC Symbol;Acc:HGNC:4752] | 193 | 5.068 | 0.1629 | Yes |
| 9 | H4C11 | H4 clustered histone 11 [Source:HGNC Symbol;Acc:HGNC:4785] | 248 | 4.803 | 0.1620 | Yes |
| 10 | ATF7IP | activating transcription factor 7 interacting protein [Source:HGNC Symbol;Acc:HGNC:20092] | 252 | 4.778 | 0.1875 | Yes |
| 11 | H2BC5 | H2B clustered histone 5 [Source:HGNC Symbol;Acc:HGNC:4747] | 279 | 4.611 | 0.2001 | Yes |
| 12 | H2BC18 | H2B clustered histone 18 [Source:HGNC Symbol;Acc:HGNC:24700] | 309 | 4.515 | 0.2106 | Yes |
| 13 | BAHD1 | bromo adjacent homology domain containing 1 [Source:HGNC Symbol;Acc:HGNC:29153] | 369 | 4.280 | 0.2042 | Yes |
| 14 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 378 | 4.234 | 0.2240 | Yes |
| 15 | CHTF8 | chromosome transmission fidelity factor 8 [Source:HGNC Symbol;Acc:HGNC:24353] | 401 | 4.154 | 0.2361 | Yes |
| 16 | H2BC15 | H2B clustered histone 15 [Source:HGNC Symbol;Acc:HGNC:4749] | 408 | 4.126 | 0.2563 | Yes |
| 17 | H2BS1 | H2B.S histone 1 [Source:HGNC Symbol;Acc:HGNC:4762] | 410 | 4.122 | 0.2791 | Yes |
| 18 | CTCF | CCCTC-binding factor [Source:HGNC Symbol;Acc:HGNC:13723] | 416 | 4.106 | 0.2998 | Yes |
| 19 | H2BU1 | H2B.U histone 1 [Source:HGNC Symbol;Acc:HGNC:20514] | 456 | 3.982 | 0.3021 | Yes |
| 20 | DDB2 | damage specific DNA binding protein 2 [Source:HGNC Symbol;Acc:HGNC:2718] | 476 | 3.934 | 0.3145 | Yes |
| 21 | H4C13 | H4 clustered histone 13 [Source:HGNC Symbol;Acc:HGNC:4791] | 477 | 3.932 | 0.3367 | Yes |
| 22 | DAXX | death domain associated protein [Source:HGNC Symbol;Acc:HGNC:2681] | 496 | 3.895 | 0.3494 | Yes |
| 23 | H2BC6 | H2B clustered histone 6 [Source:HGNC Symbol;Acc:HGNC:4753] | 517 | 3.832 | 0.3607 | Yes |
| 24 | HIRA | histone cell cycle regulator [Source:HGNC Symbol;Acc:HGNC:4916] | 518 | 3.831 | 0.3824 | Yes |
| 25 | H3C13 | H3 clustered histone 13 [Source:HGNC Symbol;Acc:HGNC:25311] | 530 | 3.801 | 0.3982 | Yes |
| 26 | H2BC13 | H2B clustered histone 13 [Source:HGNC Symbol;Acc:HGNC:4748] | 563 | 3.725 | 0.4026 | Yes |
| 27 | H2BC11 | H2B clustered histone 11 [Source:HGNC Symbol;Acc:HGNC:4761] | 582 | 3.688 | 0.4142 | Yes |
| 28 | HAT1 | histone acetyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:4821] | 590 | 3.669 | 0.4313 | Yes |
| 29 | SHPRH | SNF2 histone linker PHD RING helicase [Source:HGNC Symbol;Acc:HGNC:19336] | 603 | 3.648 | 0.4457 | Yes |
| 30 | CHAF1B | chromatin assembly factor 1 subunit B [Source:HGNC Symbol;Acc:HGNC:1911] | 622 | 3.617 | 0.4568 | Yes |
| 31 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 635 | 3.590 | 0.4709 | Yes |
| 32 | H1-4 | "H1.4 linker histone, cluster member [Source:HGNC Symbol;Acc:HGNC:4718]" | 682 | 3.514 | 0.4669 | Yes |
| 33 | H1-10 | H1.10 linker histone [Source:HGNC Symbol;Acc:HGNC:4722] | 702 | 3.481 | 0.4767 | Yes |
| 34 | H2BC4 | H2B clustered histone 4 [Source:HGNC Symbol;Acc:HGNC:4757] | 730 | 3.433 | 0.4822 | Yes |
| 35 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 740 | 3.419 | 0.4968 | Yes |
| 36 | RECQL | RecQ like helicase [Source:HGNC Symbol;Acc:HGNC:9948] | 796 | 3.320 | 0.4871 | Yes |
| 37 | LIN54 | lin-54 DREAM MuvB core complex component [Source:HGNC Symbol;Acc:HGNC:25397] | 805 | 3.309 | 0.5016 | Yes |
| 38 | FAM172BP | "family with sequence similarity 172 member B, pseudogene [Source:HGNC Symbol;Acc:HGNC:34336]" | 1168 | -3.804 | 0.3354 | No |
| 39 | SMARCA1 | "SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 [Source:HGNC Symbol;Acc:HGNC:11097]" | 1350 | -4.193 | 0.2652 | No |
| 40 | TOP1MT | DNA topoisomerase I mitochondrial [Source:HGNC Symbol;Acc:HGNC:29787] | 1521 | -4.644 | 0.2033 | No |
| 41 | PADI4 | peptidyl arginine deiminase 4 [Source:HGNC Symbol;Acc:HGNC:18368] | 1622 | -4.950 | 0.1795 | No |
Table: GSEA details [plain text format]