![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 2183_Read1.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 76394599 |
| Filtered Sequences | 0 |
| Sequence length | 75 |
| %GC | 43 |
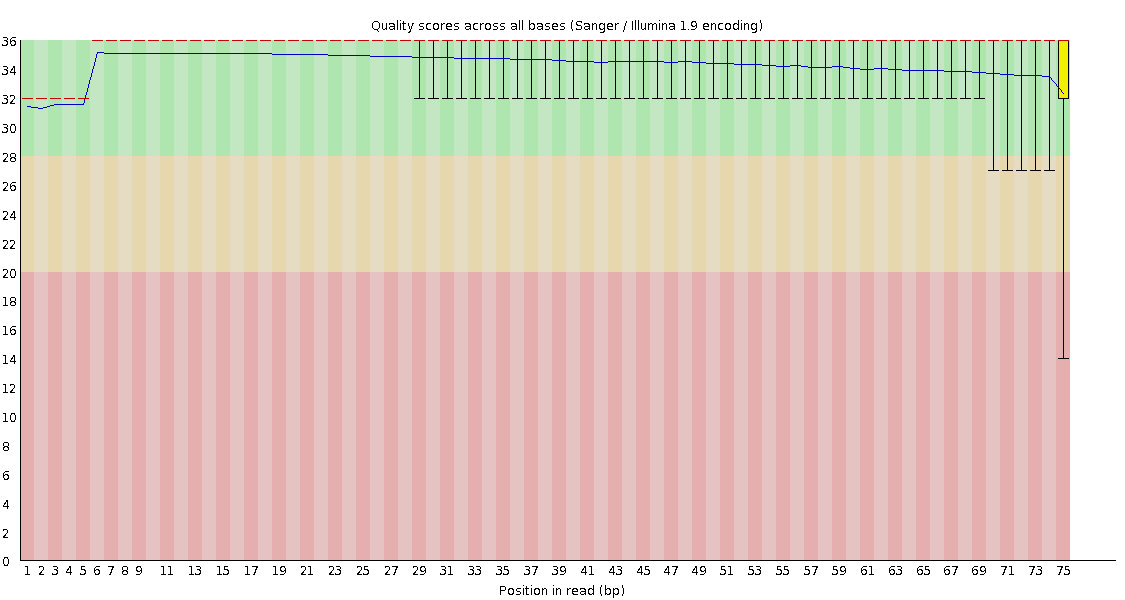
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
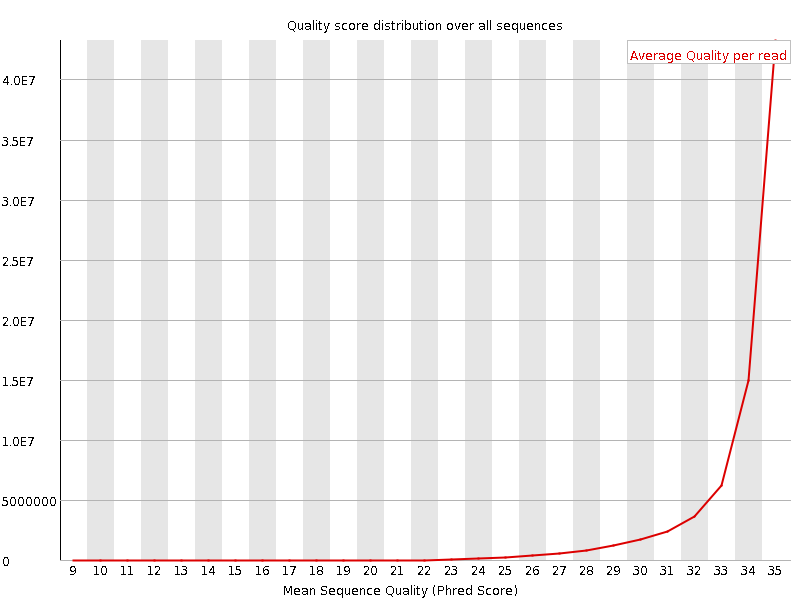
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
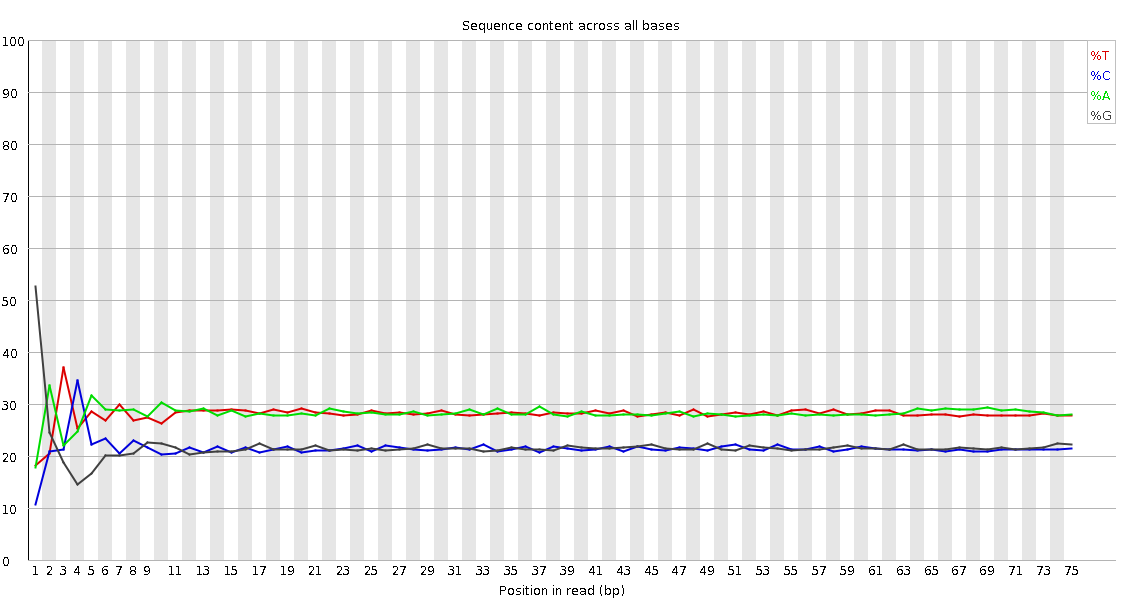
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
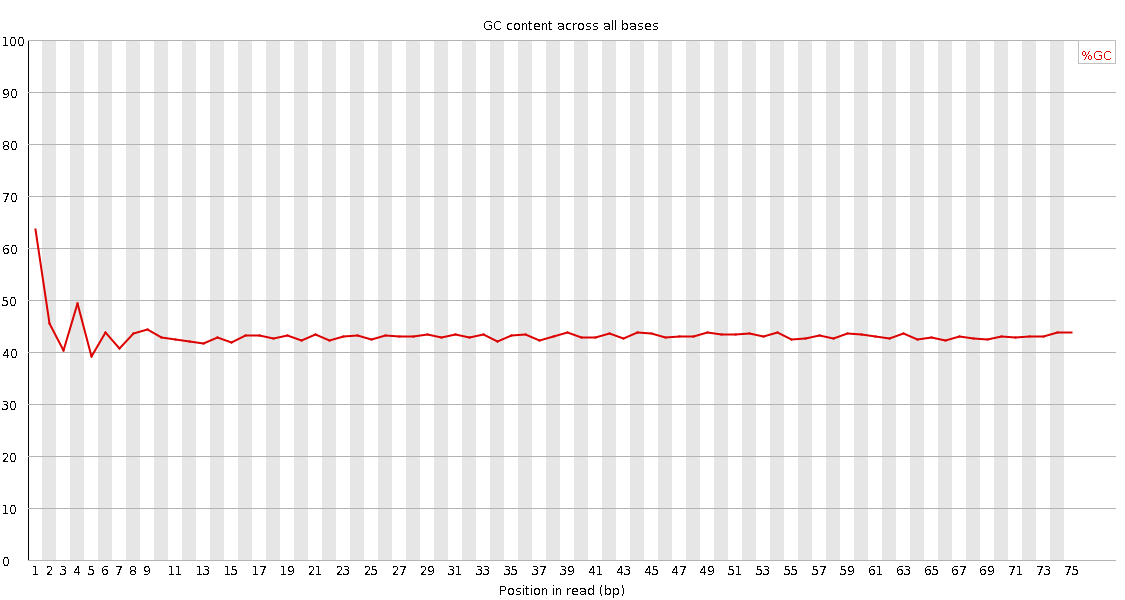
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
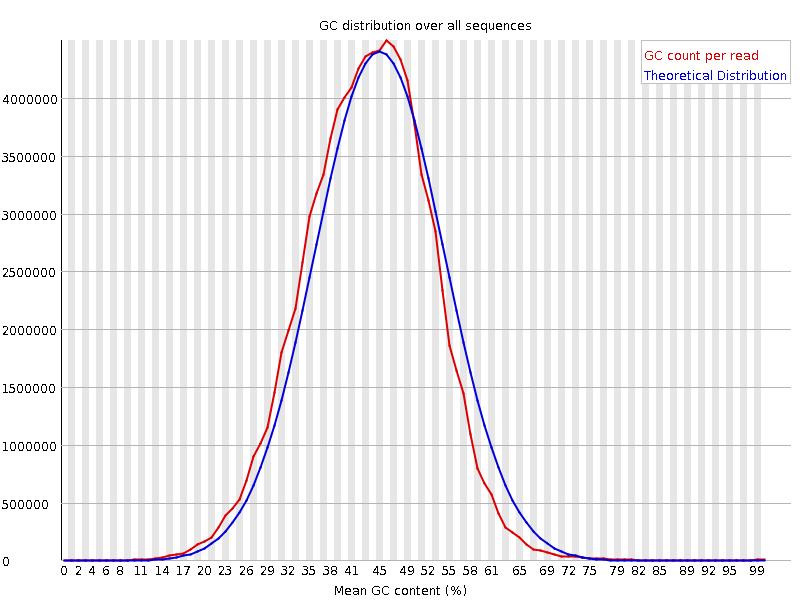
![[OK]](Icons/tick.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
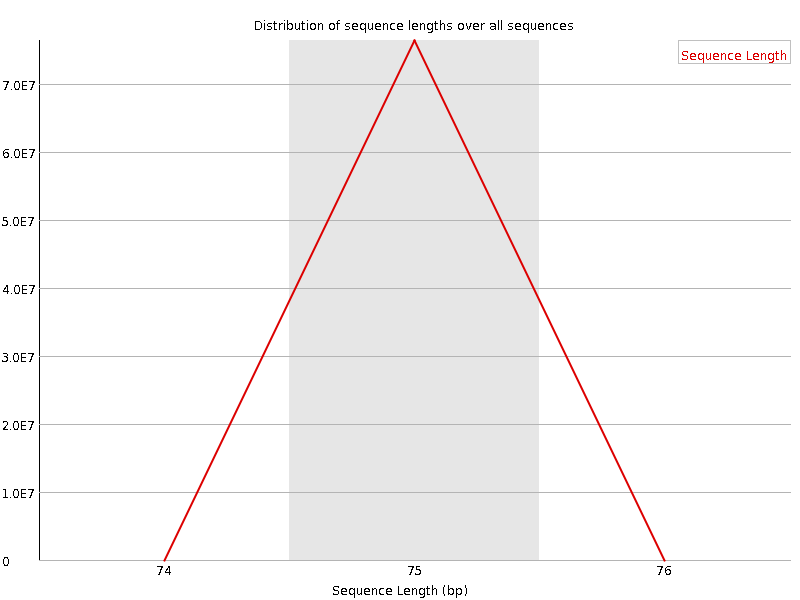
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
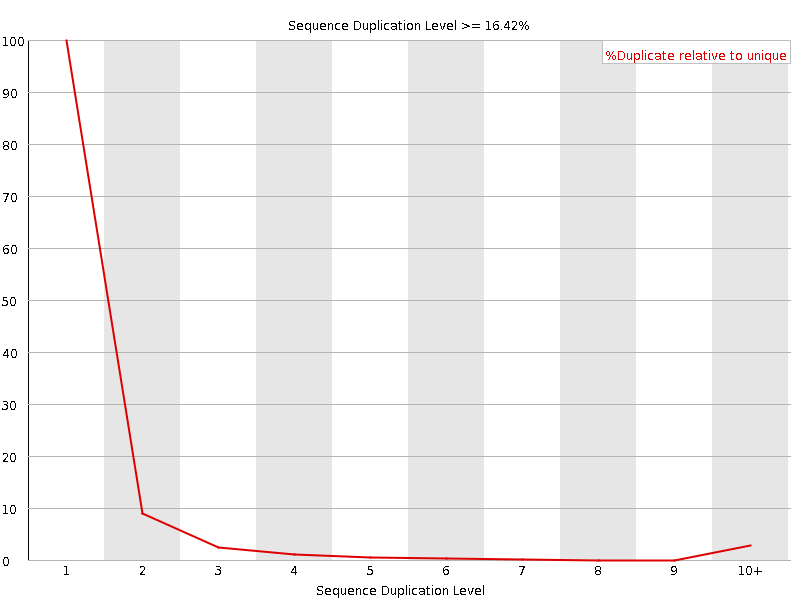
![[OK]](Icons/tick.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACATCACGATCTCGTATGCCGTCTTCTGCTTGAAAAAAAAAAGG | 162937 | 0.21328340240387936 | TruSeq Adapter, Index 1 (100% over 63bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACATCACGATCTCGTATGCCGTCTTCTGCTTGAAAAAAAAAGGG | 129470 | 0.16947533162651984 | TruSeq Adapter, Index 1 (100% over 63bp) |
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
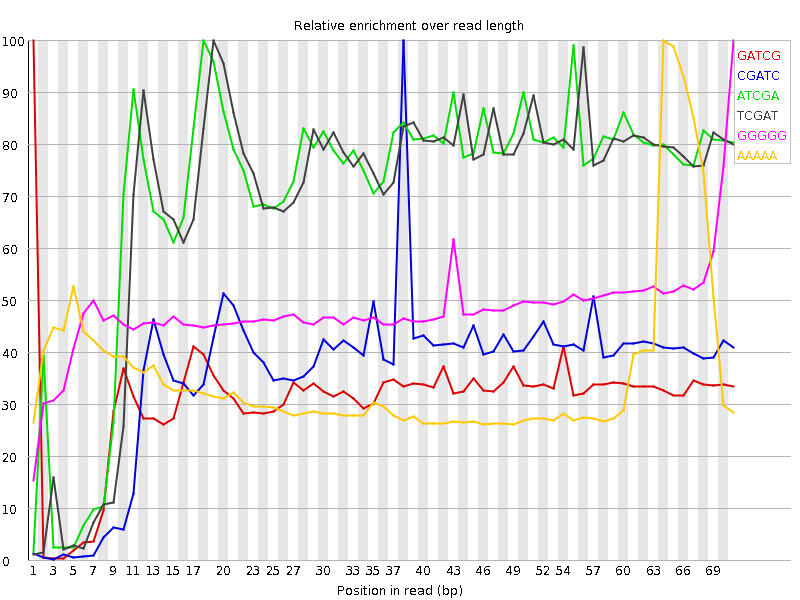
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GATCG | 24444430 | 5.4625783 | 17.667568 | 1 |
| CGATC | 22884725 | 5.1841497 | 14.449842 | 38 |
| ATCGA | 23068160 | 3.9810243 | 5.6287537 | 18 |
| TCGAT | 22591580 | 3.912492 | 5.6705813 | 19 |
| GGGGG | 7461835 | 2.7485023 | 5.707178 | 71 |
| AAAAA | 26372680 | 2.6682432 | 7.423335 | 64 |
| GATCC | 11014395 | 2.495126 | 49.323586 | 1 |
| CACAC | 10389370 | 2.3774436 | 12.784096 | 12 |
| TCCAG | 10441005 | 2.3652341 | 12.382505 | 25 |
| TCTCT | 13424325 | 2.365039 | 6.6083407 | 3 |
| GATCT | 13202195 | 2.2864041 | 42.61706 | 1 |
| TCCTG | 9827705 | 2.2341325 | 6.783183 | 3 |
| CACAG | 9788310 | 2.209605 | 5.447734 | 4 |
| CTCCA | 9245870 | 2.1232135 | 12.3032 | 24 |
| CTTCT | 12039440 | 2.121056 | 9.372442 | 54 |
| TTCTG | 12005590 | 2.086485 | 9.200255 | 55 |
| GGAAG | 9304105 | 2.0438704 | 11.864265 | 5 |
| TCCCA | 8863245 | 2.035348 | 5.8111186 | 3 |
| CTGCT | 8776435 | 1.9951472 | 11.038078 | 57 |
| GATCA | 11496840 | 1.9840857 | 39.717243 | 1 |
| TCTTC | 10938460 | 1.9270902 | 9.278755 | 53 |
| TCCTC | 8351785 | 1.924643 | 5.1335874 | 3 |
| TCTGC | 8181710 | 1.8599484 | 11.5017185 | 56 |
| TCTCC | 8023250 | 1.8489331 | 5.945966 | 3 |
| TCCCT | 8014895 | 1.8470078 | 6.7619424 | 3 |
| GAAGA | 10850800 | 1.8407917 | 9.735605 | 6 |
| TCTTT | 13606645 | 1.8326179 | 5.8541465 | 3 |
| TCCCC | 6039065 | 1.8203948 | 5.854217 | 3 |
| TCCTT | 10284655 | 1.8119056 | 5.3645334 | 3 |
| TCTGA | 10268350 | 1.7783102 | 9.505273 | 18 |
| ATCCA | 10115125 | 1.7695678 | 14.682854 | 2 |
| GAAAA | 13319260 | 1.7449665 | 7.37355 | 63 |
| TCACA | 9465915 | 1.6559932 | 9.502023 | 30 |
| AAGAG | 9683365 | 1.6427414 | 9.663781 | 7 |
| CTGAA | 9503055 | 1.640005 | 9.352885 | 19 |
| TCTCA | 9285225 | 1.6300968 | 5.1866174 | 3 |
| AGAGC | 7164025 | 1.5953277 | 11.9381075 | 8 |
| CCAGT | 7020795 | 1.5904431 | 11.564666 | 26 |
| TCCAC | 6865355 | 1.5765542 | 5.54774 | 3 |
| GAGCA | 7068635 | 1.5740858 | 11.686893 | 9 |
| TCCAT | 8953425 | 1.5718465 | 6.284271 | 3 |
| CCATC | 6801650 | 1.561925 | 5.0598345 | 4 |
| ATCTG | 8986985 | 1.5563986 | 10.801149 | 2 |
| GCACA | 6891860 | 1.5557625 | 11.683791 | 11 |
| ATCCC | 6679625 | 1.5339034 | 13.84412 | 2 |
| ATCCT | 8721755 | 1.5311749 | 13.044015 | 2 |
| GTCTG | 6803505 | 1.525722 | 11.601358 | 17 |
| CACAT | 8576125 | 1.500331 | 9.256678 | 31 |
| TGCTT | 8540725 | 1.4843166 | 8.561175 | 58 |
| AGCAC | 6534690 | 1.4751354 | 11.58414 | 10 |
| ATCTC | 8375415 | 1.4703721 | 12.246467 | 2 |
| TGAAA | 11042570 | 1.4517844 | 7.7201223 | 62 |
| CAGTC | 6253040 | 1.4165212 | 11.477155 | 27 |
| ATCTT | 10311145 | 1.383894 | 11.681231 | 2 |
| ATCAG | 7908925 | 1.3648955 | 9.447553 | 2 |
| CTTGA | 7619035 | 1.3194922 | 8.389687 | 60 |
| GCTTG | 5707895 | 1.2800257 | 10.486721 | 59 |
| ACTCC | 5524480 | 1.268637 | 11.497288 | 23 |
| TCAAG | 7348475 | 1.2681749 | 5.4773107 | 3 |
| CATCA | 7240970 | 1.2667553 | 9.033939 | 33 |
| GTCTT | 7270665 | 1.2635893 | 8.575434 | 52 |
| AACTC | 7098995 | 1.2419178 | 9.105618 | 22 |
| AGTCA | 7152030 | 1.2342731 | 8.970068 | 28 |
| ATGCC | 5396440 | 1.2224728 | 11.109101 | 47 |
| GAACT | 7075815 | 1.2211201 | 8.934551 | 21 |
| ATCAC | 6767890 | 1.1839935 | 9.1821 | 2 |
| TTGAA | 8709290 | 1.1490519 | 6.7086263 | 61 |
| ATCAA | 8554830 | 1.1401379 | 10.696623 | 2 |
| GTCAC | 5030765 | 1.1396353 | 11.237154 | 29 |
| ATCAT | 7773135 | 1.0396022 | 7.8748326 | 2 |
| ACATC | 5923095 | 1.0362027 | 8.790101 | 32 |
| TGAAC | 5998050 | 1.0351231 | 8.726947 | 20 |
| GTATG | 4998355 | 0.85392535 | 8.339999 | 45 |
| ATCTA | 6375835 | 0.85272306 | 5.2913237 | 2 |
| TATGC | 4768005 | 0.8257404 | 8.417182 | 46 |
| CGTCT | 2564170 | 0.58291286 | 10.637799 | 16 |
| CACGT | 2401540 | 0.5440285 | 10.782266 | 14 |
| TCACG | 2163495 | 0.49010342 | 10.502583 | 35 |
| TGCCG | 1655425 | 0.48559842 | 13.301227 | 48 |
| ACACG | 2120495 | 0.47867876 | 10.66204 | 13 |
| CCGTC | 1535405 | 0.45656726 | 13.30606 | 50 |
| TCTCG | 1933805 | 0.43961194 | 9.430673 | 41 |
| GCCGT | 1447755 | 0.42468098 | 13.195329 | 49 |
| CGGAA | 1867185 | 0.4157959 | 10.890266 | 4 |
| CACGA | 1781445 | 0.40214187 | 10.181672 | 36 |
| ACGTC | 1652190 | 0.3742759 | 10.609592 | 15 |
| CTCGT | 1603430 | 0.36450776 | 9.614112 | 42 |
| TCGGA | 1548620 | 0.34606895 | 11.048992 | 3 |
| ATCGG | 1499150 | 0.3350139 | 11.955654 | 2 |
| ACGAT | 1613020 | 0.27836952 | 7.541288 | 37 |
| CGTAT | 1395775 | 0.2417254 | 7.869985 | 44 |
| TCGTA | 1311625 | 0.22715199 | 7.8034286 | 43 |