| Sequence |
Count |
Percentage |
Possible Source |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTAT |
25788115 |
51.22905682935428 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| GAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTATGCCGT |
1060390 |
2.106504471973969 |
TruSeq Adapter, Index 1 (96% over 30bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATATCGTAT |
906763 |
1.8013186794674902 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGAACTCGTAT |
522755 |
1.0384723971809922 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGAGCTCGTAT |
488006 |
0.9694422064996169 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| NATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTAT |
476574 |
0.946732110097721 |
TruSeq Adapter, Index 6 (97% over 34bp) |
| CGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTATGCC |
430528 |
0.8552600055734296 |
TruSeq Adapter, Index 1 (96% over 32bp) |
| GATCGGAAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTA |
347258 |
0.6898410301198017 |
TruSeq Adapter, Index 6 (96% over 30bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGACCTCGTAT |
316818 |
0.6293708294135638 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAACGATCTCGTATG |
254725 |
0.5060207580452185 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACATCTCGTATG |
252017 |
0.5006412145658331 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| GATCGGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTA |
200169 |
0.3976432196178363 |
TruSeq Adapter, Index 1 (96% over 31bp) |
| GAATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTA |
198436 |
0.3942005501755265 |
TruSeq Adapter, Index 6 (97% over 34bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAAGATCTCGTATG |
190246 |
0.37793080826409126 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| ATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTATG |
181114 |
0.3597897480522199 |
TruSeq Adapter, Index 1 (97% over 34bp) |
| GATCGGAAGGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTA |
161073 |
0.3199775505373147 |
Illumina Multiplexing PCR Primer 2.01 (96% over 27bp) |
| GATCGGAAGAAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTA |
133161 |
0.26452931656515594 |
TruSeq Adapter, Index 6 (96% over 26bp) |
| GATCGGAAGAGCACACGTCTGAACTCAGTCACGTGAAACGATCTCGTATG |
121322 |
0.24101069941137304 |
Illumina Multiplexing PCR Primer 2.01 (96% over 26bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGAAATCGTAT |
109299 |
0.21712655936238823 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| GATCGGAAGAGTACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTAT |
101485 |
0.2016037555411483 |
TruSeq Adapter, Index 6 (97% over 34bp) |
| TGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTATGCC |
99347 |
0.1973565384218994 |
TruSeq Adapter, Index 6 (96% over 31bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAATGATCTCGTAT |
99210 |
0.19708438278797186 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| GGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTATGCCG |
98735 |
0.19614077748785808 |
TruSeq Adapter, Index 1 (96% over 31bp) |
| GATCGGAAGACACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTATG |
96411 |
0.19152406439845937 |
TruSeq Adapter, Index 1 (95% over 24bp) |
| GATCGGAAGAGCACAACGTCTGAACTCCAGTCACGTGAAACGATCTCGTA |
94032 |
0.18679809174799486 |
TruSeq Adapter, Index 1 (95% over 21bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCATGTGAAACGATCTCGTAT |
93909 |
0.18655374763870222 |
TruSeq Adapter, Index 6 (97% over 34bp) |
| GATCGGAAGAGCACACGTCTGAACTCCATCACGTGAAACGATCTCGTATG |
93328 |
0.18539956936635255 |
Illumina Multiplexing PCR Primer 2.01 (96% over 28bp) |
| GATCGGAAGAGCACACGTCTGAAACTCCAGTCACGTGAAACGATCTCGTA |
91078 |
0.1809298600500242 |
Illumina Multiplexing PCR Primer 2.01 (95% over 23bp) |
| GATCGGAAGAGCACATGTCTGAACTCCAGTCACGTGAAACGATCTCGTAT |
89602 |
0.17799773073851277 |
TruSeq Adapter, Index 6 (97% over 34bp) |
| GATCGGAAGAGCAACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTA |
86949 |
0.17272744682019317 |
TruSeq Adapter, Index 1 (95% over 23bp) |
| GGATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTA |
85089 |
0.16903248711869504 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| GATCGGAAGAGGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTA |
83828 |
0.16652746336407726 |
TruSeq Adapter, Index 1 (96% over 25bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGCCAATATCTCGTATGC |
81943 |
0.16278284022573106 |
TruSeq Adapter, Index 6 (100% over 50bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGAGATCGTAT |
79203 |
0.15733972754718006 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATTCGTATG |
74061 |
0.1471249518562643 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| GATCGGAAAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTATG |
72969 |
0.14495565293473964 |
TruSeq Adapter, Index 1 (96% over 26bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAAACGATCTCGTA |
72909 |
0.1448364606863042 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| GATCGGAAGAGCATACGTCTGAACTCCAGTCACGTGAAACGATCTCGTAT |
71390 |
0.1418189102634141 |
TruSeq Adapter, Index 6 (97% over 34bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGACACGATCTCGTAT |
70474 |
0.13999924193729996 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| GATCGGAAGAGCACACGGTCTGAACTCCAGTCACGTGAAACGATCTCGTA |
69403 |
0.13787166030272766 |
No Hit |
| GATCGGAAGAGCACACGTCTGAACTCTAGTCACGTGAAACGATCTCGTAT |
67450 |
0.13399195261615462 |
TruSeq Adapter, Index 6 (97% over 34bp) |
| AGATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTA |
66888 |
0.1328755185558095 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| GATTGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTAT |
66529 |
0.1321623516026709 |
TruSeq Adapter, Index 6 (97% over 34bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGGTCACGTGAAACGATCTCGTA |
62889 |
0.12493135519758858 |
Illumina Multiplexing PCR Primer 2.01 (96% over 29bp) |
| GATCGGAAGAGCACACGTCTGAACTTCAGTCACGTGAAACGATCTCGTAT |
62657 |
0.12447047850363827 |
TruSeq Adapter, Index 6 (97% over 34bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTTACGTGAAACGATCTCGTAT |
61888 |
0.12294283118619093 |
TruSeq Adapter, Index 6 (97% over 34bp) |
| GATCGGAAGAGCACACGTTTGAACTCCAGTCACGTGAAACGATCTCGTAT |
60060 |
0.11931144068385838 |
TruSeq Adapter, Index 6 (97% over 34bp) |
| GATGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTATG |
59702 |
0.11860026026819369 |
TruSeq Adapter, Index 6 (96% over 31bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAAGTCACGTGAAACGATCTCGTA |
59412 |
0.1180241644007558 |
Illumina Multiplexing PCR Primer 2.01 (96% over 28bp) |
| GATCGGAAGAGCACACGTCTGAACTCCCAGTCACGTGAAACGATCTCGTA |
58691 |
0.11659187088205682 |
Illumina Multiplexing PCR Primer 2.01 (96% over 27bp) |
| GATCGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTATG |
57671 |
0.11456560265865462 |
TruSeq Adapter, Index 1 (96% over 30bp) |
| GATCGGAAGAGCACACGTCTGAATTCCAGTCACGTGAAACGATCTCGTAT |
56339 |
0.11191953474338824 |
TruSeq Adapter, Index 6 (97% over 34bp) |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGACATCGTAT |
56196 |
0.11163545988461714 |
TruSeq Adapter, Index 1 (97% over 35bp) |
| GATCGGAAGAGCACACGTCTGGAACTCCAGTCACGTGAAACGATCTCGTA |
54023 |
0.1073187139537809 |
Illumina Multiplexing PCR Primer 2.01 (95% over 23bp) |
| AATCGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTAT |
53029 |
0.10534409570470073 |
TruSeq Adapter, Index 6 (97% over 34bp) |
| GATCGGAAGAGCACACGTCTGAATCCAGTCACGTGAAACGATCTCGTATG |
52079 |
0.10345688510447319 |
Illumina Multiplexing PCR Primer 2.01 (95% over 23bp) |
| GACGGAAGAGCACACGTCTGAACTCCAGTCACGTGAAACGATCTCGTATG |
52036 |
0.10337146399309449 |
TruSeq Adapter, Index 1 (96% over 32bp) |
| GATCGGAAGAGCACACGTCTAACTCCAGTCACGTGAAACGATCTCGTATG |
51601 |
0.10250732019193767 |
Illumina Multiplexing PCR Primer 2.01 (95% over 22bp) |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[FAIL]](Icons/error.png) Per base sequence quality
Per base sequence quality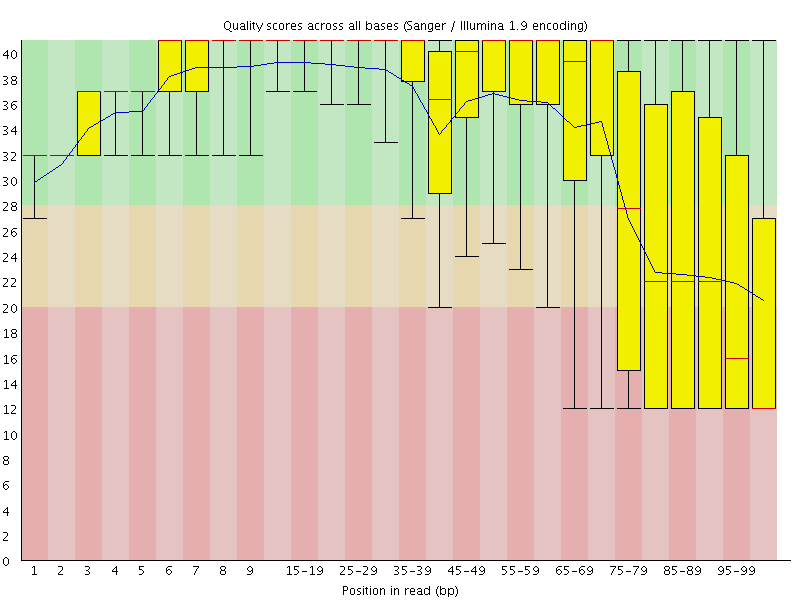
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores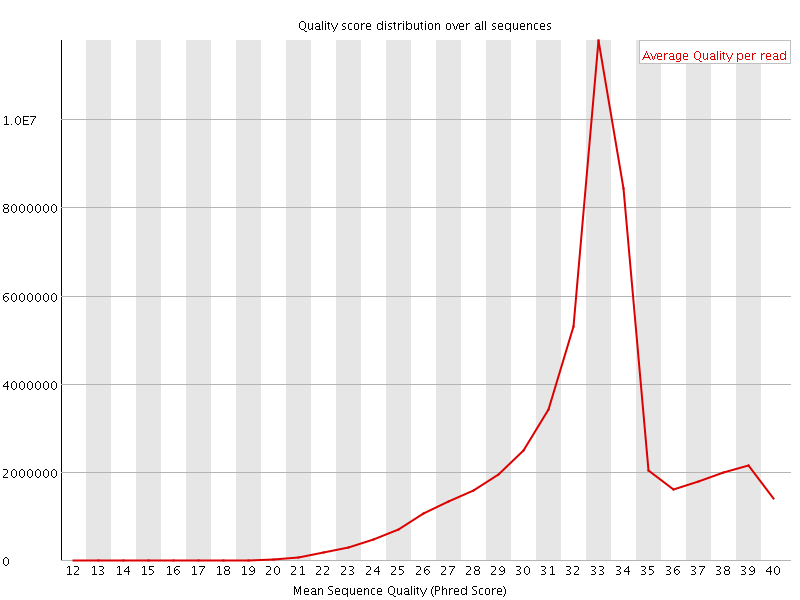
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content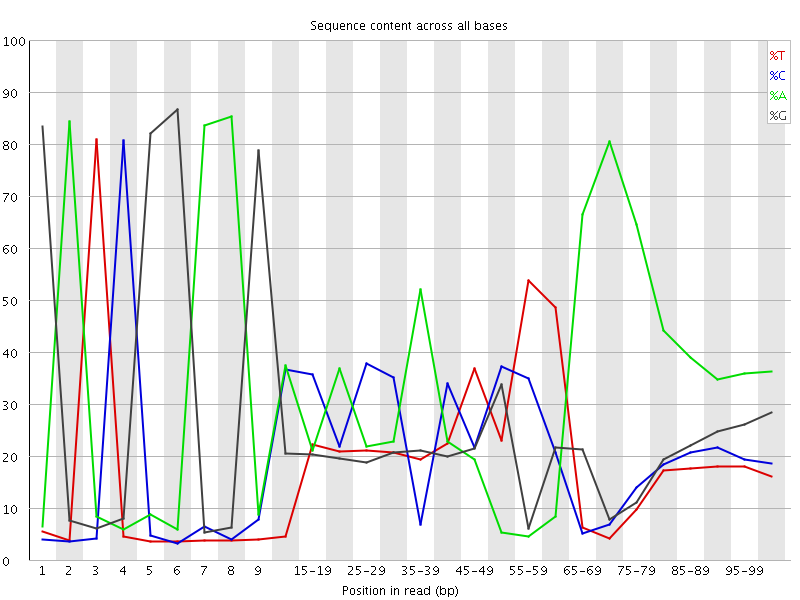
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content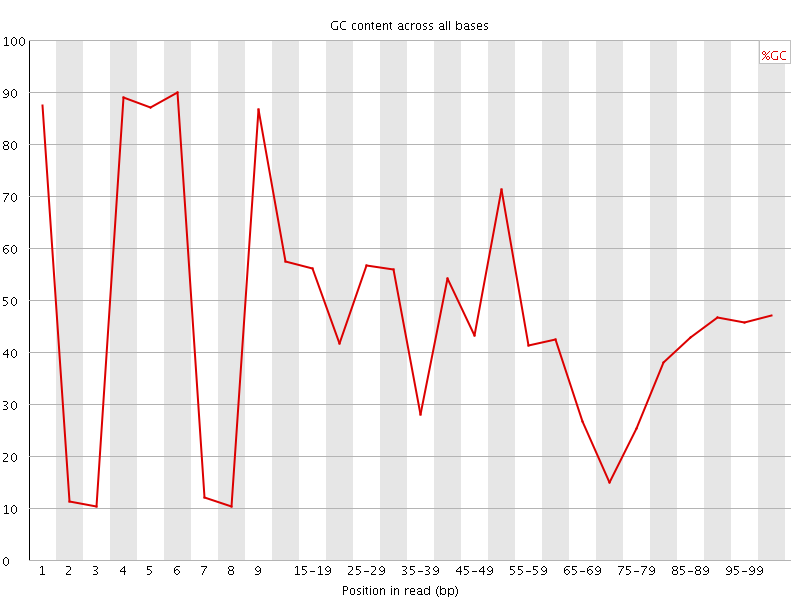
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content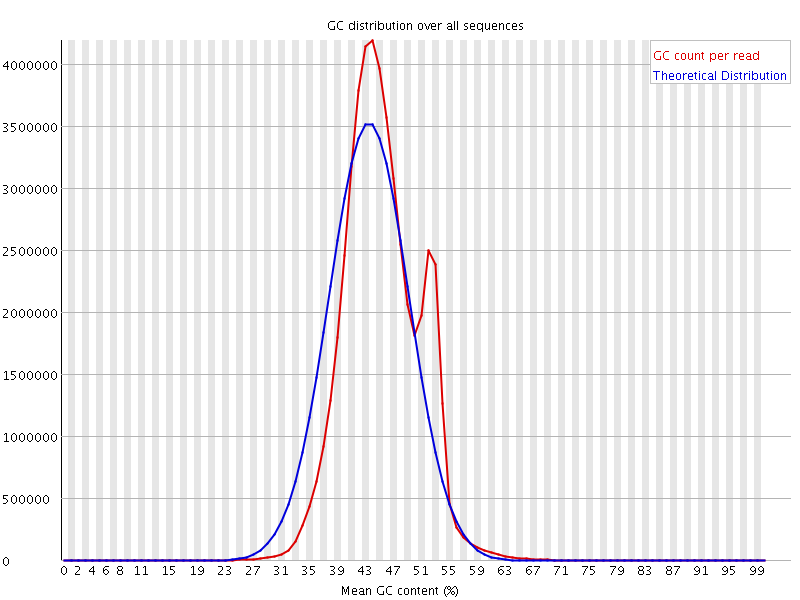
![[OK]](Icons/tick.png) Per base N content
Per base N content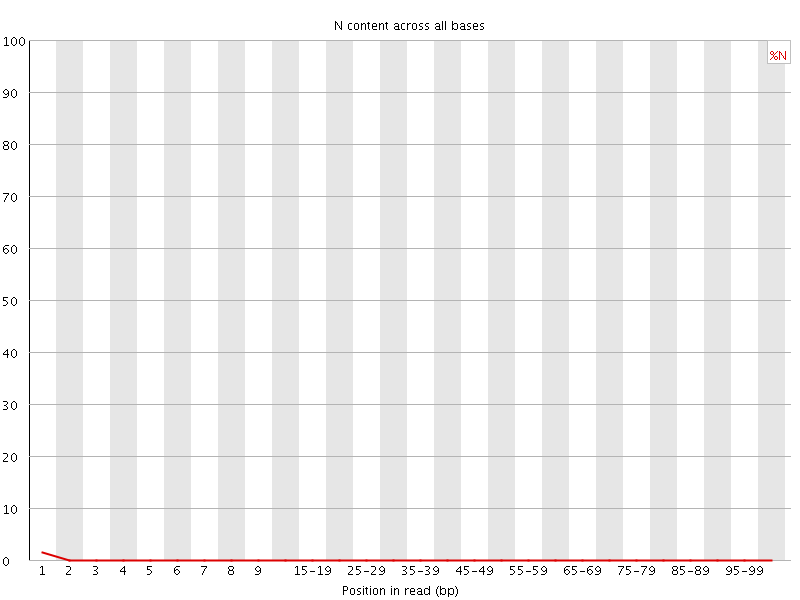
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution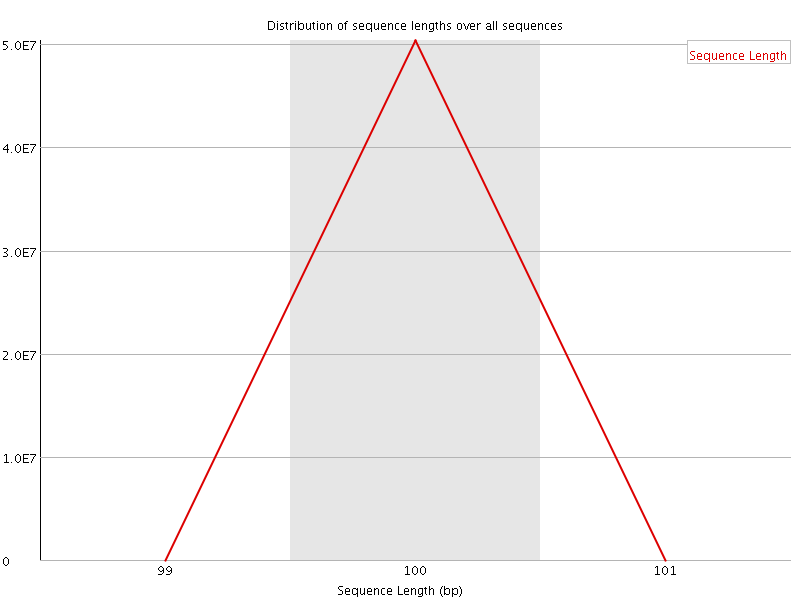
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels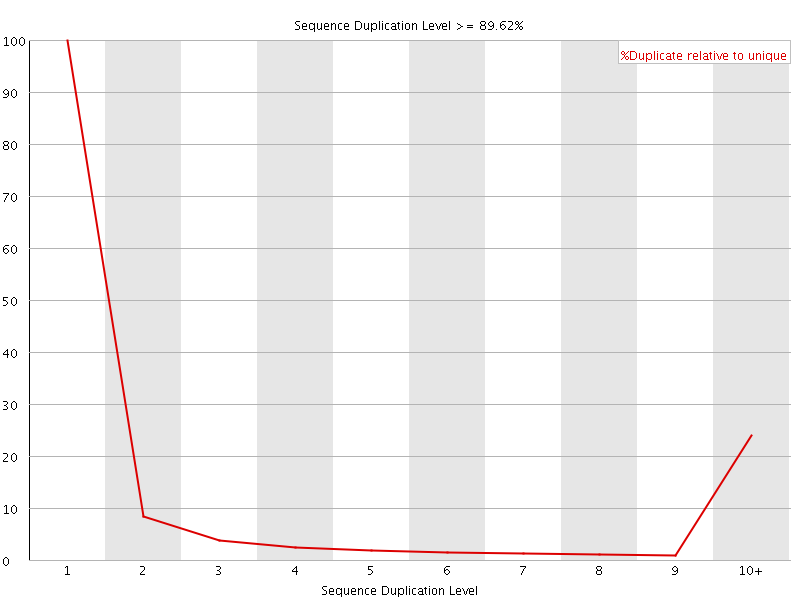
![[FAIL]](Icons/error.png) Overrepresented sequences
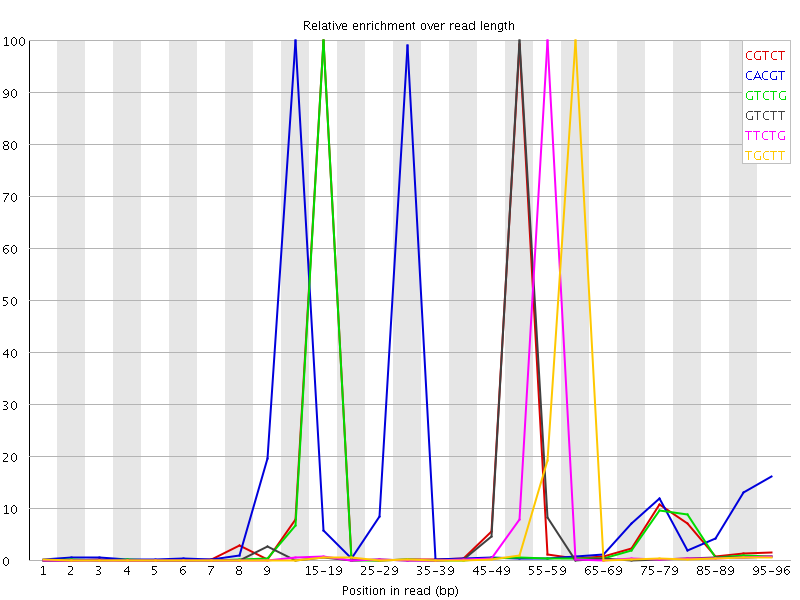
Overrepresented sequences![[FAIL]](Icons/error.png) Kmer Content
Kmer Content